In a previous post, I discussed different platforms for organism engineering that were presented at SB5.0.
Here I’ll try to give a high-level picture of Ginkgo’s pipeline for organism engineering. If you’ve checked out our webpage, you’ll see that we have several different organism engineering projects happening at Ginkgo that span several different hosts. Our goal was to build out a pipeline that could support the engineering of all these very different organisms for very different purposes but that uses a shared pipeline. To accomplish this goal, we deliberately opted to decouple design from fabrication. Ginkgo organism engineers place requests via our CAD/CAM/LIMS software system. Those requests are then batched and run on Ginkgo’s robots.
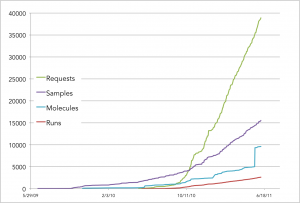
By decoupling design from fabrication and pushing construction and testing through a shared, automated pipeline, we’ve been able to achieve a level of productivity that would have been unattainable if we used conventional, manual molecular biology approaches. Below we show a plot of requests (placed either by Ginkgo organism engineers or by other pipeline processes), samples (physical objects containing DNA/strains/reagents), molecules (abstract objects corresponding to unique DNA sequences including but not limited to standardized parts), and runs (batches of multiple requests that have been completed via the Ginkgo pipe).
Hence, Ginkgo organism engineers are free to focus on design and analysis of novel organisms rather than mindless pipetting operations better done by robots than PhDs. We’re building a team of organism engineers each of whom
- cares passionately about engineering organisms
- has a strong opinion about how to engineer organisms
- understands that their opinion is just that – an opinion – and that the only way to develop general design principles for organism engineering is to design a bunch of organisms successfully
If you seek to be one of the best organism engineers on the planet and don’t want to be limited in the complexity of the organisms that you engineer by how fast you pipet, you should come talk to us. See the website for details.
