Ginkgo has just announced a commitment to buy 100 million base pairs of DNA from Twist Bioscience in 2016. By volume, this equals 10% of the global volume for gene synthesis in 2015. Here, I provide some context for how we, the customer, view an agreement like this.
Engineering biology has become a big deal – booming investment in a range of markets, an academic ecosystem, and thriving educational initiatives. These activities are powered by a distributed, expensive, and largely manual effort to construct DNA. As the field develops, the role of DNA synthesis will become increasingly important. For example, at Ginkgo we make heavy use of sequences that must be synthesized because they don’t exist in any strain collection. Additionally, as our engineering becomes more sophisticated, each nucleotide becomes a target for design so writing the DNA from scratch is necessary. Ginkgo has always outsourced DNA synthesis and has ongoing relationships with synthesis companies including Genscript and Gen9. Our agreement with Twist is a logical extension of this approach.
We believe DNA synthesis belongs in specialized facilities serving many customers. In principle, outsourcing DNA synthesis should be easy; the customer provides sequence requests, the service provider returns sequence-perfect DNA. This is an attractively simple interface. However, large-scale adoption of synthesis services depends on cost, turn time, and what sequences can be synthesized. Let’s look at each in turn.
Cost: Synthesis costs are driven by labor, and materials. The leading synthesis firms drive down these costs using automation and miniaturized reactions that support substantial economies of scale. Their custom platforms incorporate glass or silicon chips, laser-printed reagents, multiplexed sequencing, and process control software. A researcher working in a conventional lab is simply not equipped to compete with these platforms on price, which are now crossing the $0.10/bp barrier. To appreciate the progress this represents, 2 years ago the market price was $0.30/bp for gene synthesis, 10 years ago it was $1.00/bp. We expect that synthesis platforms will become increasingly differentiated from the standard wet-lab in the future. Our partnership with Twist ensures we will be on the leading edge of the ensuing falling cost curve. Falling prices don’t mean a shrinking market since an ever larger fraction of the work done by researchers at the bench, such as PCR’ing from a template, can now be replaced by synthesis.
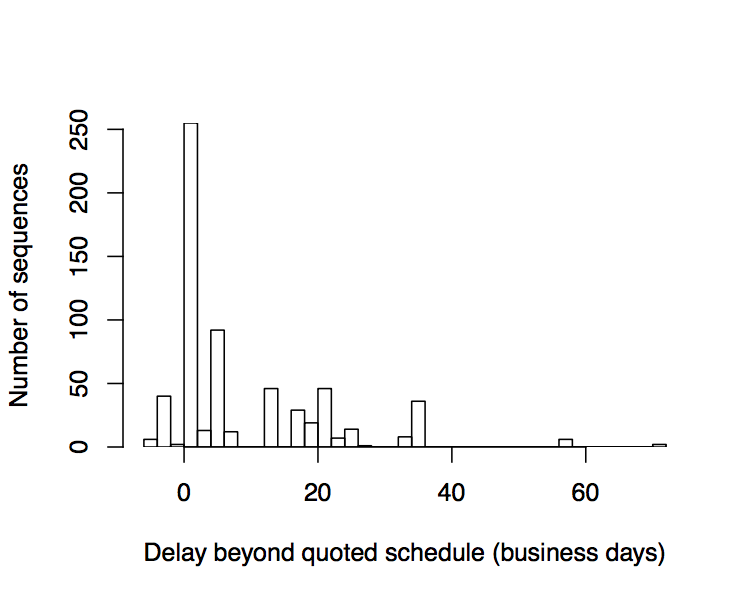
Turn time: The process time for DNA synthesis ranges from 8-20 business days and has stayed mostly constant in recent years. Those times are competitive with an expert researcher at the bench and should be sufficient to drive rapid growth of the synthesis market. However, errors in the synthesis process often extend turn time to a substantial and unpredictable degree. These uncertain delays impact the ability of a customer like Ginkgo to hit deadlines and meet our customer’s needs. Ginkgo has been tracking the difference between expected and actual turn times and some of our data is presented below. Interestingly, the delayed sequences tend to cluster around a delay of 20 business days, suggesting a second attempt was required to successfully build the sequence. While turn time will inevitably improve, we should seek ways to avoid long delays. For example, giving the provider and the customer a mechanism to cancel orders that prove hard to synthesize keeps everyone moving forward on a more predictable schedule. We have designed our agreement with Twist to provide such a mechanism that protects both side’s interests. At the same time, we must consider what makes certain DNA sequences hard to synthesize in the first place.
Sequence constraints: Repeat sequences, local regions of extreme GC percentage, and sequences constitutively expressing protein are often flagged as problematic by synthesis firms. These constraints are gradually being elucidated and loosened by ongoing process improvement. What we can do today is ensure that our designs avoid as many problematic sequences as possible. Some providers will endeavor to optimize the customer’s sequences to make them easier to synthesize but we believe that is not the right general solution. The customer needs to own the sequence design process in its entirety since they alone understand the design objective. This means integrating the synthesis constraints into the design process so all constraints can be considered together. Sharing of detailed synthesis constraints depends on trust and close relationships between customer and service provider. We think supply agreements like the one we have reached with Twist, and other companies previously, form the basis of that relationship.
Sequence length: Synthesizing long sequences (5-20kb) requires additional steps and effort and has consequently carried a price premium per base pair. This places customers in a bad spot. As sequence length (and design complexity) increases, our confidence in the design tends to go down, meaning we need to test more variants. But if the cost of each variant increases with length, the service becomes uneconomic as length increases. While falling prices in general help this problem, that is unlikely to be sufficient any time soon. A better solution is to rely on combinatorial assembly. This means synthesizing common sequences only once and then reusing that DNA in multiple longer constructs. Reusing DNA in this way spreads the synthesis cost across multiple constructs and leads to lower costs per base pair than shorter constructs. This works for the customer because sets of long sequences often involve considerable sequence reuse. Note that the 100 Mbp we will be ordering from Twist does not include combinatorial assembly.
With continued progress on the parameters discussed above, it seems plausible that all in vitro DNA synthesis, assembly, and editing will happen at specialized firms and the role of the customer will be focused on the equally interesting challenge of introducing that DNA into their organisms. Lastly, we should remember that this is a young and rapidly learning field. Only through efforts at the scale of what we are doing with Twist will we learn the rules of fast and efficient DNA synthesis, and sophisticated design of organisms. We at the Ginkgo foundry, with our friends at Twist and elsewhere, are delighted to be leading the charge.