In our last blog post, Liam and Aileen told us about their summer. Lets hear what Kevin and Vidya have been up to!
Kevin Lin, Software Engineering
My name is Kevin and I’m currently pursuing my Bachelor’s and Master’s in Computer Science at Stanford University. This summer I interned with the Decepticons, a team that works on software-automation interfaces like our Laboratory Information Management System (LIMS), a system housing data foundational to our foundry operations and codebase growth.
I first became interested in synthetic biology a little over a year ago. I took a class called “Inventing the Future” at Stanford d.school, during which Drew Endy gave a phenomenal overview of synthetic biology, its potential to solve our most pressing problems, and associated challenges like biosecurity (I highly recommend this podcast if you’re looking for a similar overview). While browsing the web to try and satiate my newfound interest, I came across this blog and was ecstatic to learn about all the ways in which software can contribute to synthetic biology. I hope this post will give yet another example of how software can make biology easier to engineer, and how someone who is relatively new to the world of biology can make an impact!
Before I begin, I want to give a huge shout out to my mentor, David Zhou, and our team’s product manager, Daniel Yang, for conducting the technical and customer research, respectively, that made this project possible.
Main Project: Improving Protein Tracking
My main project was standardizing and automating the storage of proteins in our LIMS. For context, Ginkgo uses the Design-Build-Test-Learn (DBTL) cycle to engineer biological systems to produce desired outputs. Examples of desired outputs include capping enzymes for mRNA vaccines, microbes that destroy environmental contaminants, and even the fragrance of a plant that hasn’t been seen since 1881. I worked closely with the Protein Characterization (PC) team, which is involved in the Test step of the DBTL cycle. They plan and execute experiments that provide us with insights into the proteins we engineer.
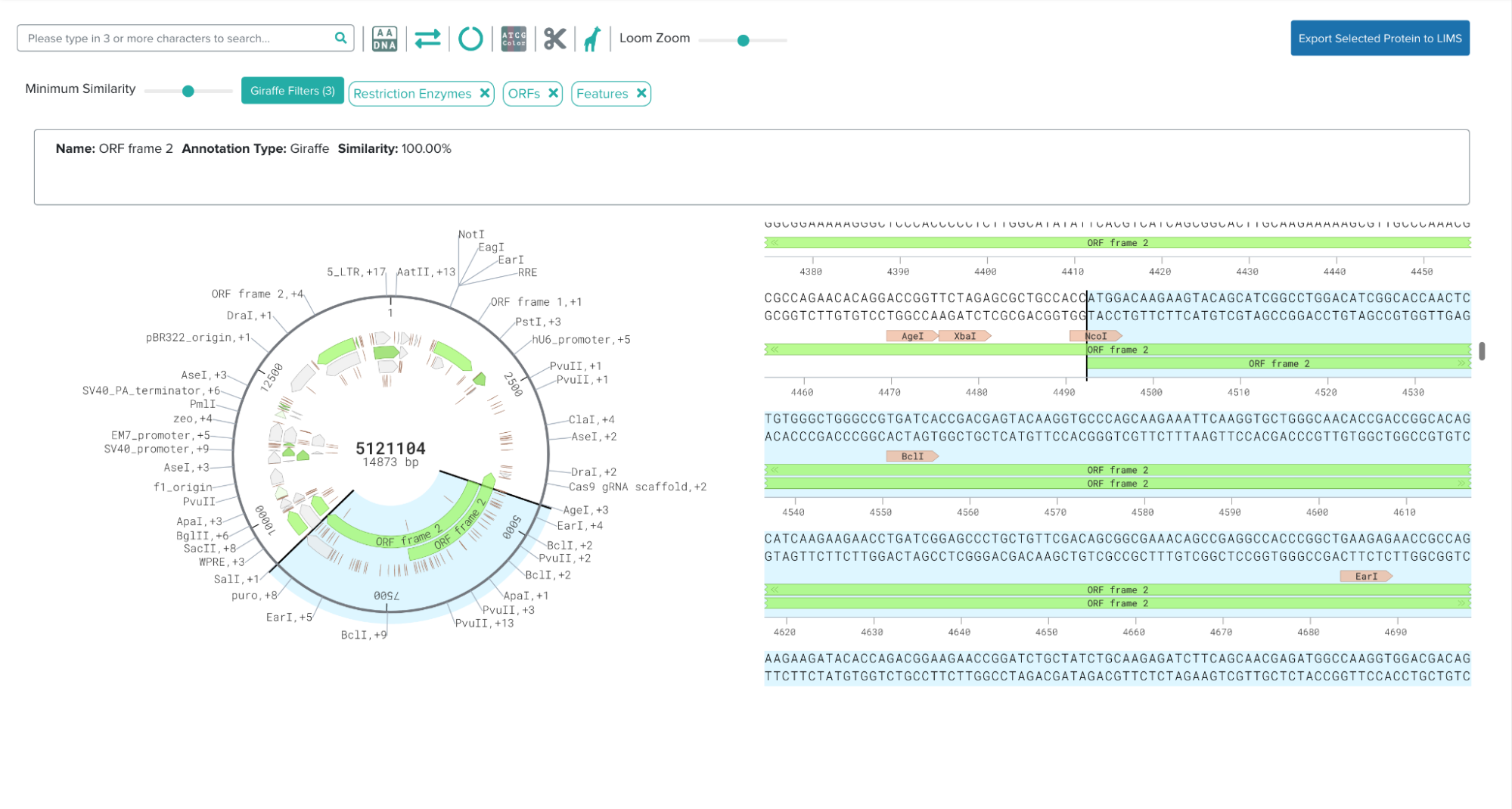
In order to successfully plan their experiments, the PC team needs to understand the work done in prior steps by gathering all the relevant data in a usable format. Part of the data gathering process involves translating designed DNA sequences into their corresponding amino acid sequences, and storing them, along with associated metadata, in our LIMS. Currently, this process is done manually, which can be time-consuming, error-prone, and inconsistent. The feature I implemented addresses these challenges by introducing a UI that automates and standardizes how we translate DNA sequences and store them in our LIMS.
Biologists can now go to our DNA sequence viewer, select an open reading frame (ORF), and export the corresponding amino acid sequence as a protein to our LIMS. Important metadata such as the DNA molecule that encodes the protein is also stored in the database.
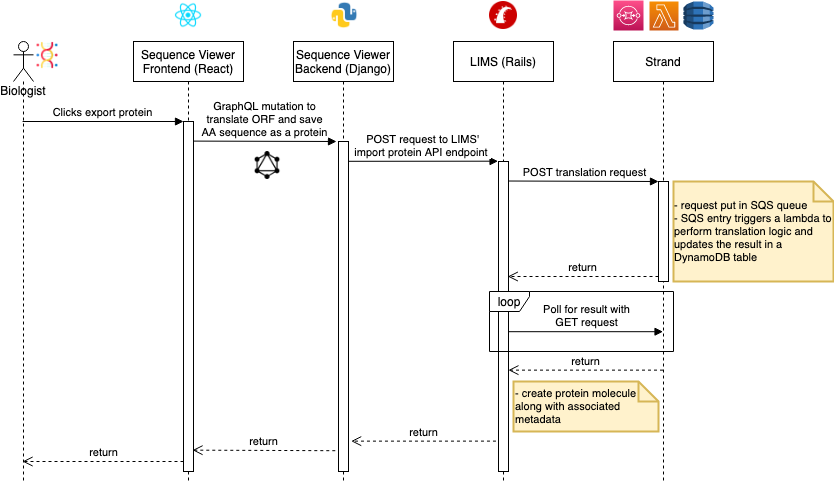
A sequence diagram for this feature is shown below. Note that Strand is a Ginkgo-specific application for bioinformatics operations hosted on AWS.
Now that this feature has been implemented, we have a framework to further standardize and automate the tracking of proteins. Potential next steps include automating the naming of proteins, organism-specific translations, and recording other important properties like relevant affinity tags and post-translational modification sites.
My Experience
I greatly enjoyed Ginkgo’s culture of growth, collaboration, and whimsy. For my project, I worked closely with Bilobans from a wide array of backgrounds – fellow software engineers, PMs, biologists, and UX designers. Each gave invaluable insights into how to make the feature better, and helped me understand a range of new topics, from what affinity tags are and why they’re important, to the design choices one can make when incorporating GraphQL into a system. Outside of my project, I reveled in learning from other teams’ office hours, weekly technical talks, monthly town halls with the founders, AMAs with senior software engineers, and textbooks I obtained with Bonusly points (best trade deal in the history of trade deals). If there’s one thing I learned from all the interdisciplinary work, it’s that anyone can help make biology easier to engineer, just like anyone can cook!
I’m going to miss the Ginkgo community and all the brilliant Bilobans that make this company what it is. This summer has been an absolute delight and I can’t wait to see what comes next!
Vidya Raghvendra, Software Engineering
Hi there! My name is Vidya, and I’m a returning software engineering intern at Ginkgo. This summer, I interned on a software team called the Impressionists, and last summer, I worked with the Decepticons (as you can tell from the team names, whimsy is a core tenet of Ginkgo’s culture!). My work last summer was focused on the back-end of the software stack, and this summer I’ve done full-stack work with a front-end focus. To learn about my perspectives and projects from last summer, check out this post and this one.
Project 1: Service Subscriber Notifications
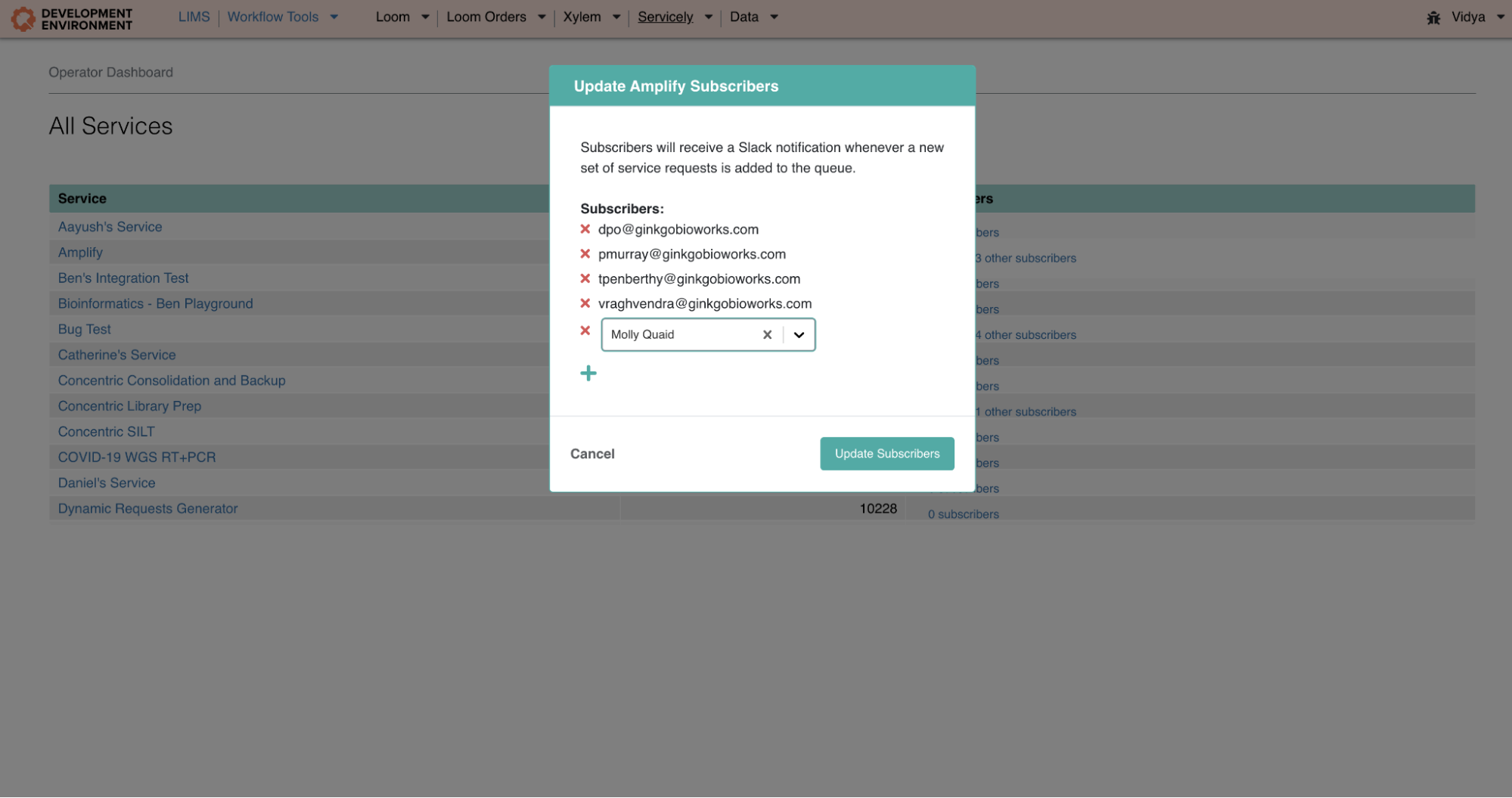
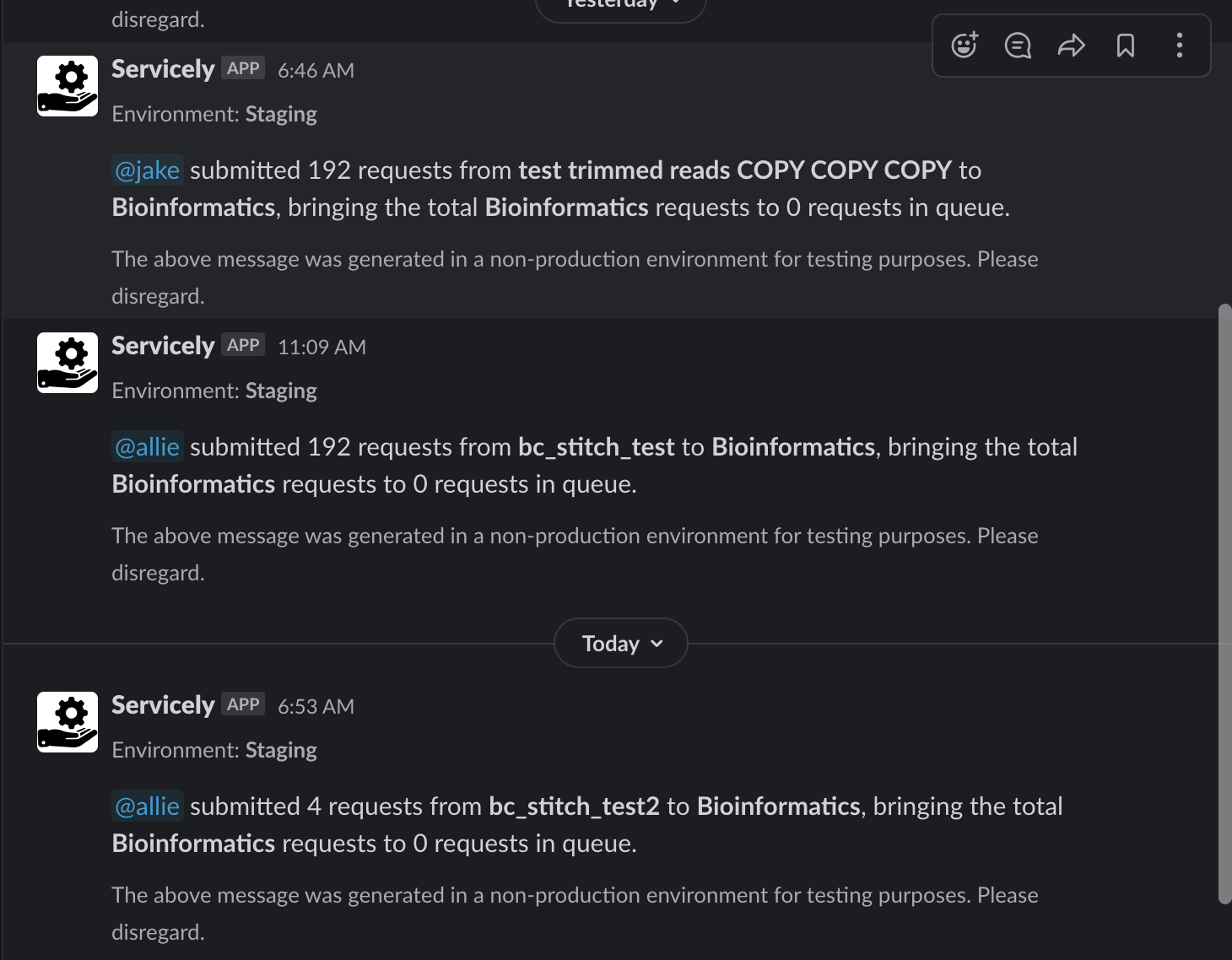
My first project focused on Servicely, one of Ginkgo’s core platforms which organizes and orchestrates the work being performed in our Foundry. On Servicely, Organism Engineers (who design and grow genetically modified organisms in Ginkgo’s labs) make requests for our services to run (e.g., for sequencing to be performed on a sample). Our Foundry operators use Servicely to execute these requests. For this project I revamped a piece of Servicely called the Operator Dashboard to enable users to add or remove subscribers to particular services– these changes would then update in the UI, and subscribers would be notified via Slack each time a new set of service requests drops into the queue. I was really eager to work with technologies I hadn’t used before, and worked with React (for the front-end) and Django (for the back-end). It was very exciting to work on a feature that would touch several users–as a result of these changes, Servicely operators no longer need to refresh queues looking for samples that they may have accidentally missed, and can avoid the potential miscommunication that comes with messaging teammates about service sample submissions.
Project 2: Simple Services
My second project also involves Servicely, as well as OrganiCK (Ginkgo’s system for notating laboratory operations). When this project is complete, Foundry operators will be able to onboard new laboratory processes (which we call “services”) using the existing user interface already available in OrganiCK. I am working on the back-end code for this project, but the rest of my scrum team will pick up the front-end after my internship ends
In addition to general streamlining and better user experience, the impact of this project includes efficiency gains from the ease of quickly onboarding laboratory processes onto a common platform, the capability for operators to more easily organize their work, and faster submission request processing.
My Ginkgo Experience (thoughts after two internships!)
Working on projects with stakeholders across the company taught me more about building for users, and it was really exciting to work with not only the engineers on my team, but also designers and product managers.
Also, although this internship was mainly remote due to COVID-19, I did have the opportunity to go into the offices in Boston in person for a week. There, I was able to meet my team and fellow interns, have lunch and an AMA with Ginkgo’s founders, and tour the Bioworks (where the magic happens!).
I think the fact that I returned to Ginkgo for a second internship attests to how much I’ve enjoyed working and learning here. Now, as Ginkgo grows and prepares to go public, I’m really glad that the culture of growth, community, and dedication to making biology easier to engineer has stayed the same and it’s still an awesome place to intern!
Conclusion
As this summer wraps up, we want to thank our Digital Tech Team interns for their contributions. What they accomplished has been impressive. Thank you, and we wish you great success in what you all will be doing next!
(Feature photo by frank mckenna on Unsplash)
