A few months ago, a number of us on the Digital Tech team had the opportunity to participate in a pilot of an introductory course in molecular and synthetic biology. Developed and taught by Natalie Kuldell and BioBuilder specifically for the “non-biologists” working at Ginkgo Bioworks, this week-long class was roughly divided evenly between classroom instruction and hands-on lab time.
In the classroom, we learned the foundations of molecular biology, from the structure of DNA and RNA, to codons, promoters, ribosomal binding sites, and primers, to protein synthesis,transcription, translation, DNA replication, and PCR. “[P]erhaps the most astonishing feature of biology is that it runs on digital code in the form of DNA, which makes it possible for us to imagine building such living machines. The code is made up of A’s, T’s ,C’s, and G’s, and we can read and write it to program cells like we program computers.” As a software engineer, this idea has always intrigued me, but until taking this course I had never fully appreciated the depth of this statement. When engineering an organism, you can componentize the A’s, T’s, C’s, and G’s into building blocks of different levels of abstraction, much like you can abstract the 1’s and 0’s of a computer program into higher level statements, functions, services, and systems.
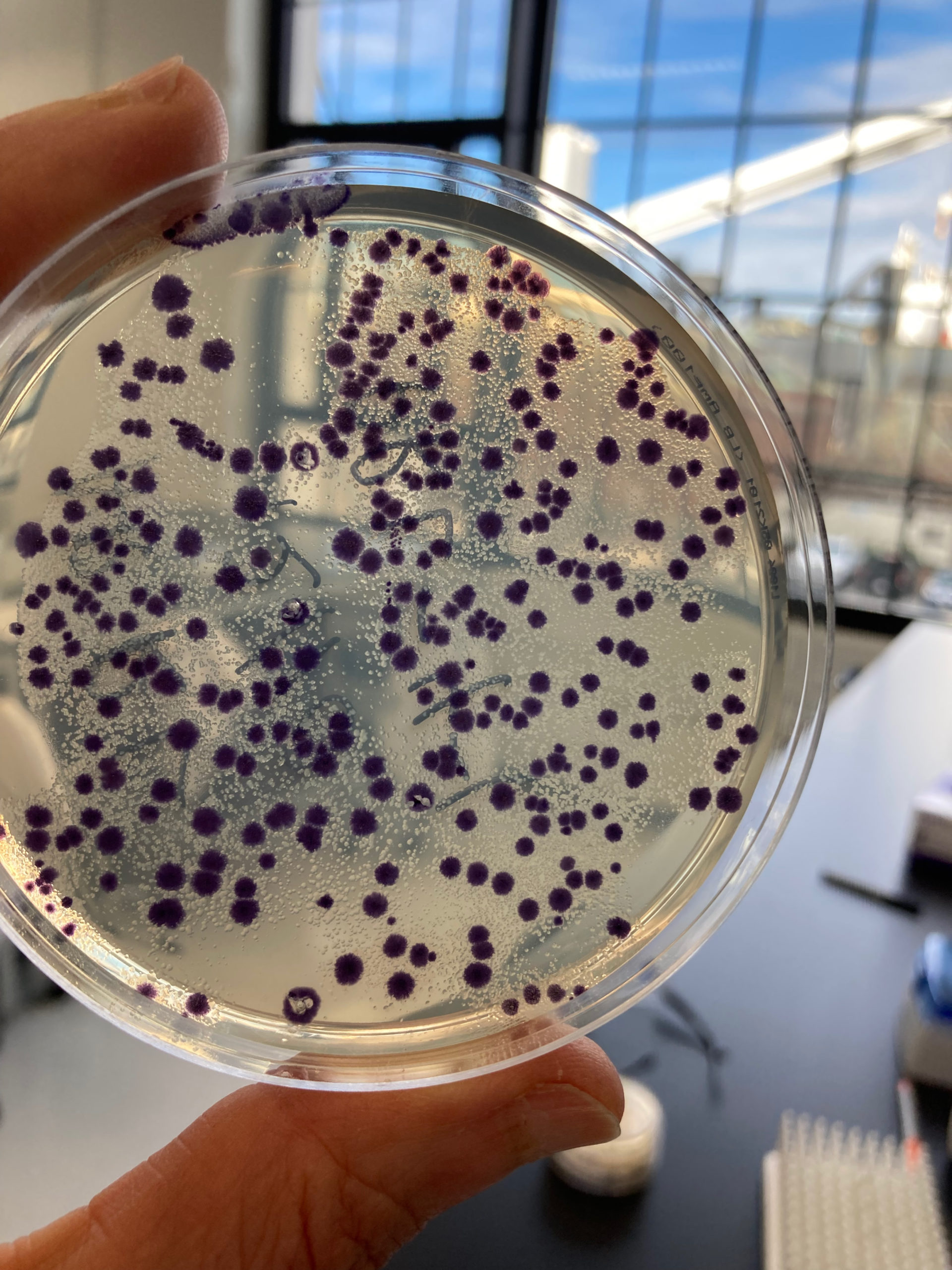
Personally, perhaps my favorite part of the course was actually engineering an organism in the laboratory. We transformed different plasmids into samples of bacteria — which is to say, we actually inserted DNA into living organisms and “programmed” them to do things for us! (In our case, we caused the bacteria to produce different colored pigments.) We later sent the samples upstairs to one of our Bioworks (BioBuilder and Ginkgo Bioworks are both located in the same building) to have them sequenced.
I very much enjoyed this course and now I more fully appreciate the work that my colleagues in the lab do each day that I help to support. Two of my colleagues from DevOps, who took the course with me, will now share their thoughts.
Dannerys Duran
I started my career at Ginkgo 4 years ago and my current role is as a DevOps Manager. I joined intending to help make biology easier to engineer. But at some point, I thought, “how do I contribute without a biology background”? Many other great contributors aren’t scientists either, and they do it, so I decided to just dive in and do my best. As the company grew, so did the non-scientific employees’ interest in learning about what happens in the lab. That’s how small sessions of BIO101 were born. Tom Knight initially hosted these sessions at Ginkgo, but they were never formalized as an official offering. However, the interest from individual contributors in Digital Tech is to learn more about what goes on in the labs and how we could bring technology closer to science. That’s how a BIO101 class of 4 half-days came about.
Having the opportunity to go to BIO101 was an eye-opener for me on how engineering biology works and how the magic happens in the lab. We learned how DNA is structured, the different types of DNA, retrieving reagents, growing overnight cultures, verifying DNA, testing it, how to analyze it, and much more. We also learned about some essential tools in the workbench and how to use them. Some of the tools included pipettes, glass tubes, cuvettes, vortex, incubators, reading the absorbance using the spectrophotometer, and of course, a lab coat, glasses, and gloves — because safety first!
The instructor was passionate about everything biology offers and was excellent at transmitting that passion through her class. She made learning about promoters, inverters, the system, and different growth phases (LOG, LAG, and stationary) easy.
Going into the lab and creating experiments on what we learned after each session, understanding the how and why of all the different components or compounds can yield a variety of results, and how to interpret them. The experience was exciting and enlightening because it better explained what the scientists we support actually do every day.
One of the class highlights was sequencing our samples using our homegrown tools, as it was very close to what the process is like for the scientists. After the experience, I decided to stick to computers! However, it provided a much more clear understanding of all the commonly used terms and practices around synthetic biology. I am able to now engage in conversations where I wouldn’t have understood the biological side of things before.
By any means, I wouldn’t say I am a biology expert, but I now have a better understanding of the practice on how to make Biology easier to engineer.
Jim Waldrop
I graduated from a top-tier university with a bachelor’s and master’s degree in Electrical Engineering and Computer Science. However, the last time I touched biology was in high school, and I’d never touched modern bioinformatics or synthetic biology at all. Despite having personal connections who did quite a bit of synthetic biology, I could never quite grasp some of the fundamentals. This class changed that.
I learned about DNA transcription and protein synthesis, binding sites, and our current understanding of how ribosomes work. I learned what metabolomics and proteomics are, as concepts, and why you can’t just code a bunch of “logic gates” into a strand of DNA without taking into account all of the other metabolic pathways in a cell or organism. We got to see how genes are encoded, and how you can have a single base pair be part of up to 6 genes at a time, theoretically, so it really drove home to me how careful you need to be when rewriting or inserting DNA into a cell to make sure that you’re not altering other genes when you do it. I learned enough to understand why we don’t just have ‘compilers for cells’ – imagine having to recompile every bit of software on your computer, including the operating system, every time you wrote a small program! That’s the problem you’d need to solve if you wanted to “just make a compiler, but for DNA”!
These are concepts that I’d been struggling to grasp for years, but just being able to sit down and have someone talk through them in a short course was what I needed.
My team maintains quite a large amount of computational resources for our Foundry scientists to use to “do bioinformatics”. Previously, we were stuck just handwaving away the specifics of what our users were doing, and just provisioning our systems for the use case of “They’re doing bioinformatics! It’s science! They need compute resources!” Now I can use the knowledge I got from this class to extrapolate what our users in the Ginkgo Foundry are using these computational resources for. I already have a background in classical computer science; I understand algorithmic run times, and I’m familiar with maintaining enterprise-scale computing systems. But to properly maintain such a system, you really do want to understand what it’s being used for. This insight will help us across our entire computing infrastructure, and that will allow our scientific users in the Foundry to get their jobs done better and faster.


(Feature photo of a lapel pin representing a promoter, ribosomal binding site, and coding region by Natalie Kuldell)
