Once again, the Digital Technology Team welcomed a number of interns this summer. This is the first of a two-part series where our interns tell you about their summer in their own words. We will begin with Ariel, Yasmine, and Evan!
Ariel Fuchs, Software Engineering
Introduction
Hello! I am Ariel and I am pursuing a Bachelor’s Degree in Computer Science and Brain and Cognitive Science at MIT. This past summer I have been working on the Impressionists team as a full stack software engineer intern. Before I dive into my summer project, I want to give a BIG shoutout to Ben Hyman and Taylor Penberthy for being amazing mentors and enabling me to grow significantly from when I first started at Ginkgo!
Project
The overarching goal of my project was to create a personalized settings page for Ginkgo services. On this page, users can tailor services so when they are working with different tools, all of their preferences will be pre-selected. Since this project will keep expanding after my internship, my specific summer project was to create the foundation and build out the necessary components for Servicely Users. Servicely is a unified applications portal for Ginkgo’s Foundry services, and is designed with two different personas in mind: requesters and operators. Requesters are users who make requests to Foundry services. Operators are users who carry out Foundry service runs in the lab. I worked on the front end in React, and implemented the backend portion using a Django framework.
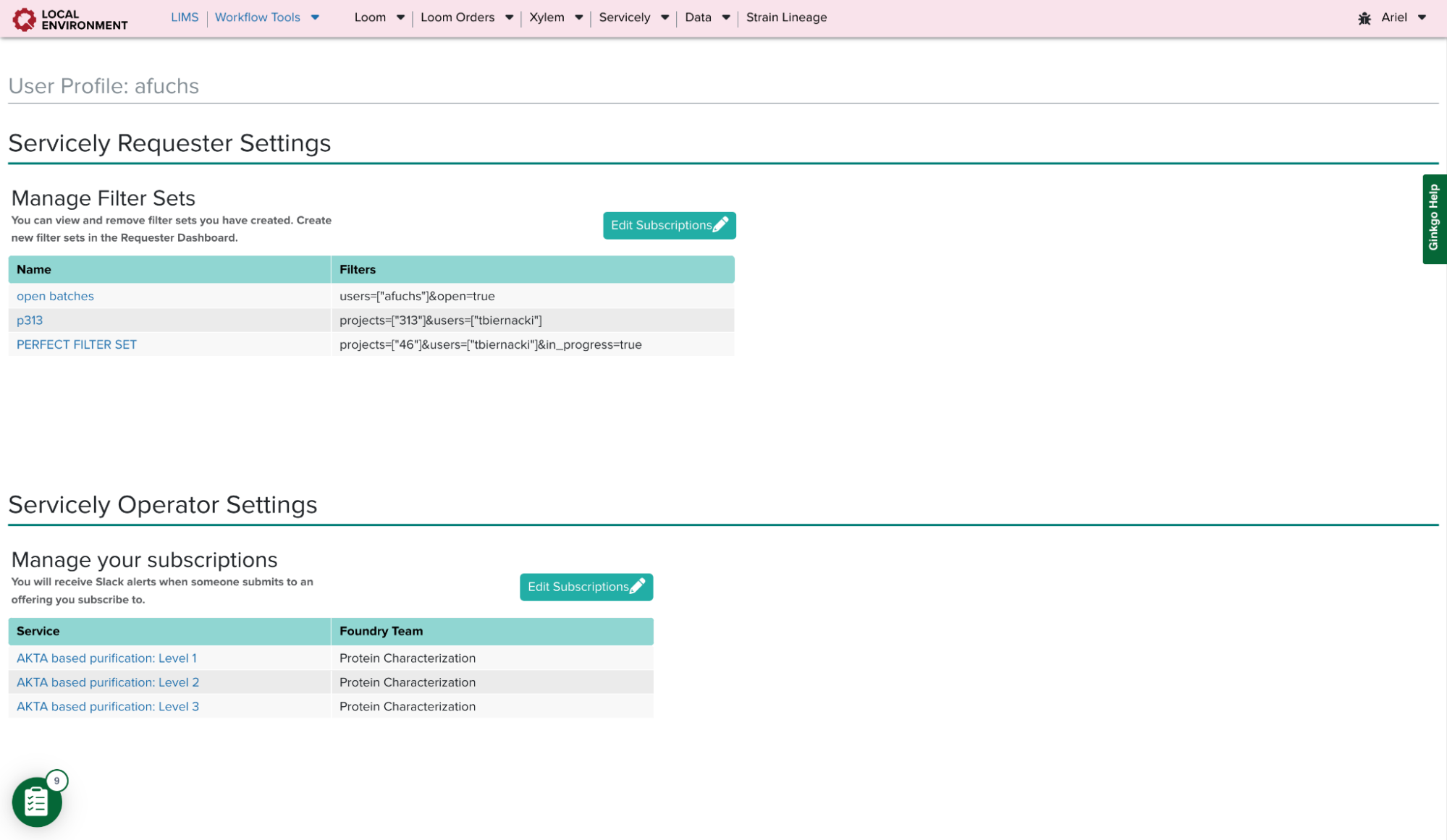
Operator Settings Page Component
On Servicely’s Operator Dashboard, there is a list of all services and a list of all users who subscribe to each one. Being subscribed to a service allows for necessary slack notifications when requesters submit offerings. However, this page is not designed to specifically show services relevant to a user. Therefore, in the settings page, I created a table where users could manage all of their subscriptions. Operators can now see all services they are subscribed to, and they can easily add or remove themselves from multiple services at a time.
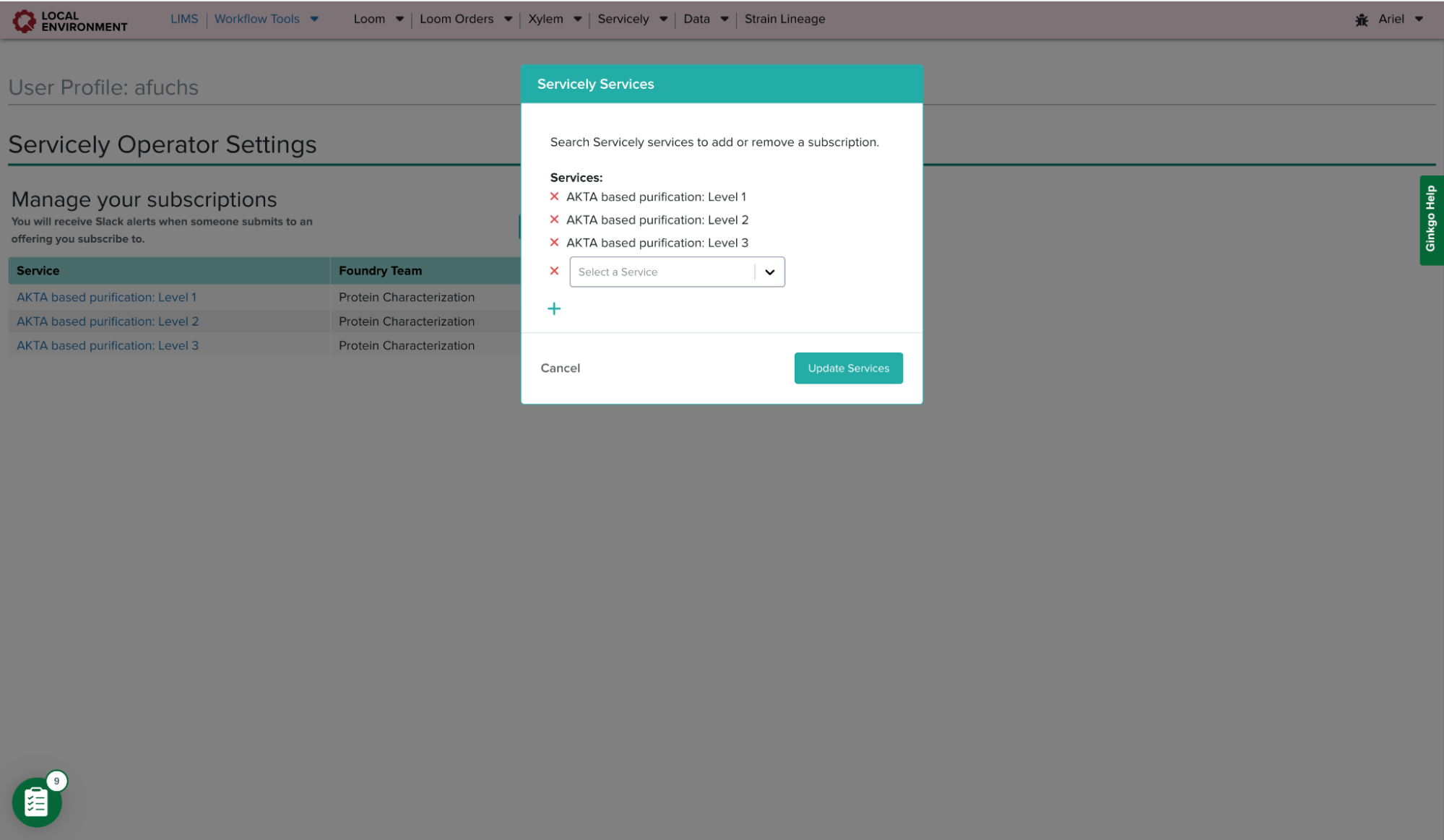
Requester Settings Page Component
On Servicely’s Requester Dashboard, there is a display of all batches created. This table displays the status of each batch, the project and service it is associated with, and the Foundry team and users involved with the request. Similar to the Operator Dashboard, this page is not designed to show which batches are relevant to a user unless multiple filters are applied. Therefore, to make the Servicely requester experience more efficient, I introduced the notion of custom filter sets. These filter sets are a collection of filters that are commonly applied together by the requester. Users can create unlimited filter sets and apply them easily in the existing filter panel. Requesters can manage these filter sets in the settings page by setting their default filter set or removing any filter sets they have created.
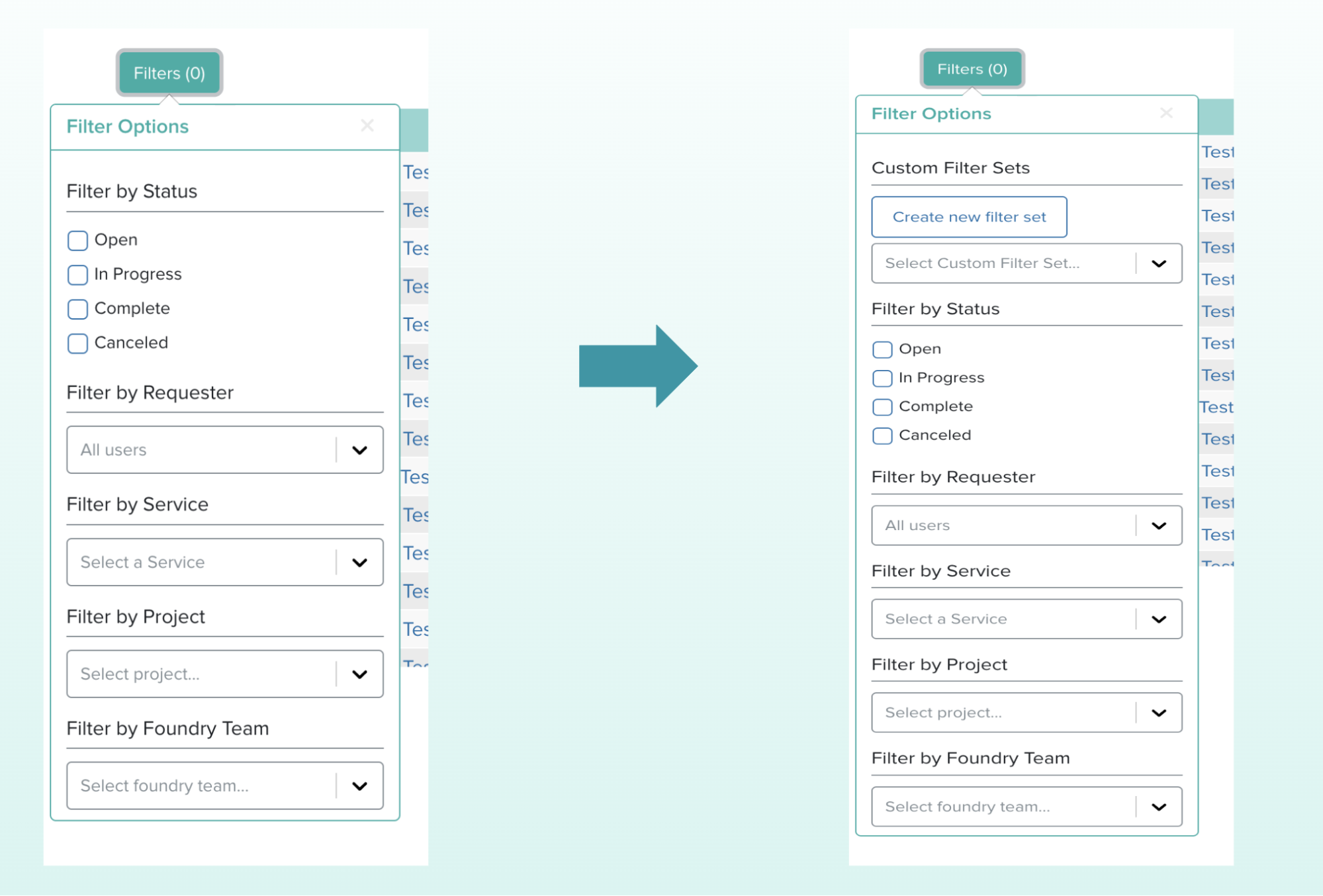
Ginkgo Experience
Besides working on my summer project and taking on additional software tickets, I was able to meet with many senior leaders at Ginkgo and expand my technical background. It was extremely exciting to meet the founders and learn about their experiences first hand. In addition, everyone I worked with was very knowledgeable and I learned more with every interaction I had with them. It was amazing to work on a project that would have high visibility and be used by many users across Ginkgo’s platform. My last highlight of the summer was the intern cohort. I enjoyed getting close to them through amusement parks, movies, and many ~Ginkgo sponsored~ meals:) Overall this was a very impactful summer and I am very grateful to everyone who helped me succeed!
Yasmine Karni, Software Engineering
Introduction
Hello! My name is Yasmine, and I just graduated with my Bachelor’s in Computer Science and Biology from Northeastern University, where I will also be completing my Master’s in Bioinformatics. This summer I had the pleasure of being the Software Engineering Intern with Ginkgo’s Decepticons team, which has a main focus of building and maintaining LIMS and OrganiCK services. LIMS is the Laboratory Information Management System, which tracks information on all of the samples in the Ginkgo Foundry. OrganiCK, aka the Organism Construction Kit, is a sister-service to LIMS meant to help users plan their experiments, note progress, and interact with samples in LIMS while running a wet lab workflow.
Main Project: Cre Lox Content Update Step
My project’s focus was to build a new content update step on OrganiCK. Content update steps in OrganiCK allow users to track the physical steps that occur on their benchtop as part of a workflow, and they also get LIMS to modify the relevant sample information accordingly, so that the LIMS samples stay up to date with the physical state of the biological samples.
The new content update step that I implemented was a Cre-Lox Recombination step. Previously, this step was being handled semi-manually by a Python package written by a Ginkgo Organism Engineer, and did not integrate with the OrganiCK and LIMS workflows. The existing solution was not easy to use and required biologists to run Python code themselves, while not being able to maintain and calculate sequence annotations or integrate into our strain lineage tracking system. By integrating the cre lox content update step into LIMS and OrganiCK, this crucial annotation and strain lineage tracking functionality is added, and the process for tracking these recombination events is streamlined. Before I dive into the specifics of my project, let me describe what a cre-lox recombination consists of.
Cre-Lox Recombination System
The cre-lox system is a site-specific recombinase system. A set of cre recombinase enzymes recognize, cut, and recombine a pair of target lox sites, resulting in a targeted insertion, deletion, or inversion of the molecule. The lox sites adjacent to the affected flanked DNA are recombined during this process. See the diagrams below for an example of each:
Inversion

When the targeted DNA region is located between two lox sites with inverted spacers facing opposite direction, in the presence of cre recombinase, the targeted DNA will be inverted.
Deletion
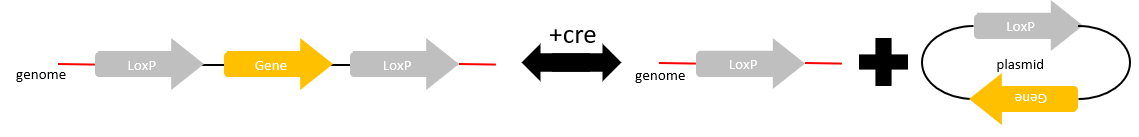
When the targeted DNA region is located between two lox sites with congruent spacers facing the same direction, in the presence of cre recombinase, the targeted DNA will be removed along with a lox site into a circular plasmid.
Insertion
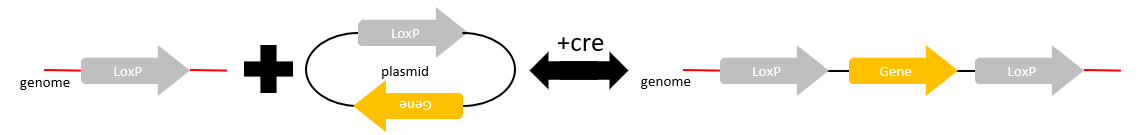
Finally, when one linear DNA molecule or genome and a circular plasmid each have a lox site, in the presence of cre recombinase, the plasmid DNA will be integrated into the linear lox site. An insertion is just the opposite of a deletion!
Building the Content Update Step
As you can see, a cre-lox recombination is a complex biological process, requiring an equally complicated bioinformatic computation to track the reorganization of sequences. This is where Strand comes in. Strand is a bioinformatics microservice that helps LIMS calculate the sequences that would result from genetic editing operations, like for instance: the annealing of DNA (joining complementary strands), digesting DNA (cutting DNA by way of restriction enzymes), or PCR (polymerase chain reaction).
So, the first step to building a new content update step was to first build a new Strand endpoint to handle the bioinformatic process of a cre-lox recombination. This involved creating a new AWS lambda function for cre-lox, and surfacing an endpoint which could be used to spin up lambdas on command to perform the calculation in a scalable way. Of course I also had to add the cre-lox operation to the core bioinformatic library which powers Strand. Unit and integration testing allowed me to ensure that the entire concert of the AWS infrastructure from the beginning of a Strand request through receiving a response from the endpoint worked together reliably.
Now that the new Strand endpoint was implemented, the next step in creating the content update step was to define the behavior we want on OrganiCK and LIMS. If you look at the diagram below, you can see the dataflow of a sample in the lab undergoing a cre lox recombination, and how that content update step is being propagated through the various levels of infrastructure. I needed to create a new OrganiCK step which would call into LIMS to update the samples; in order for LIMS to update the samples, it needed to call into the new cre lox endpoint on Strand to calculate the new sequences.
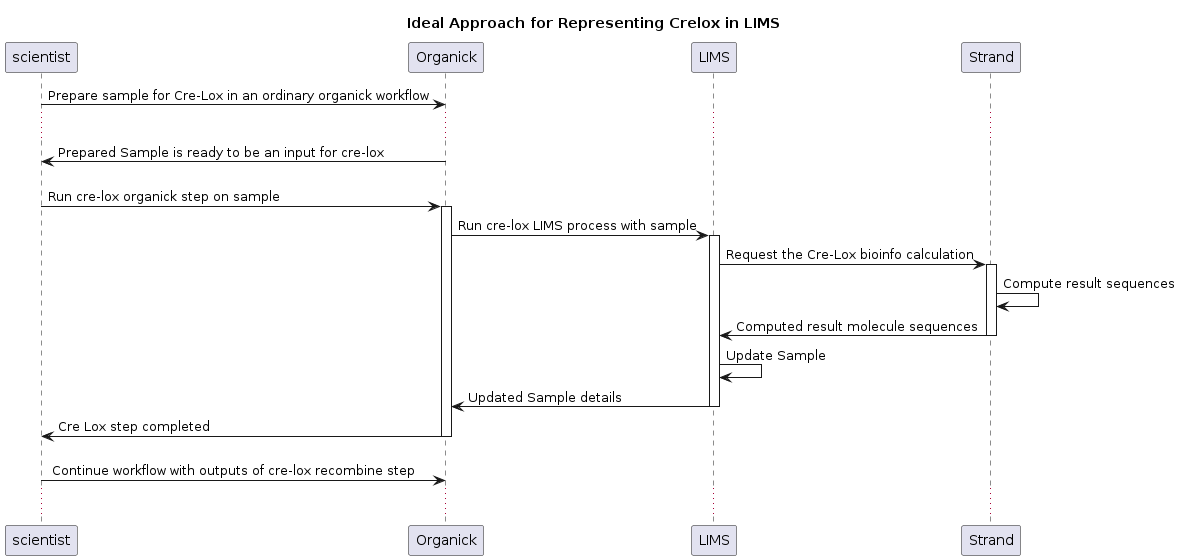
Adding Annotations
Once the first draft of the cre lox content update step was completed and live on OrganiCK, the final step to fully integrating the product molecules with Ginkgo’s LIMS was to make sure they maintain annotations once they undergo a cre-lox operation. Annotations provide context to molecules, such as where a start codon or a promoter is located – or in the case of cre-lox, where a lox site might be! Propagating annotations for the cre lox operation through Strand and finally onto LIMS was the final step to completing my project. See below for an example of annotations on LIMS:
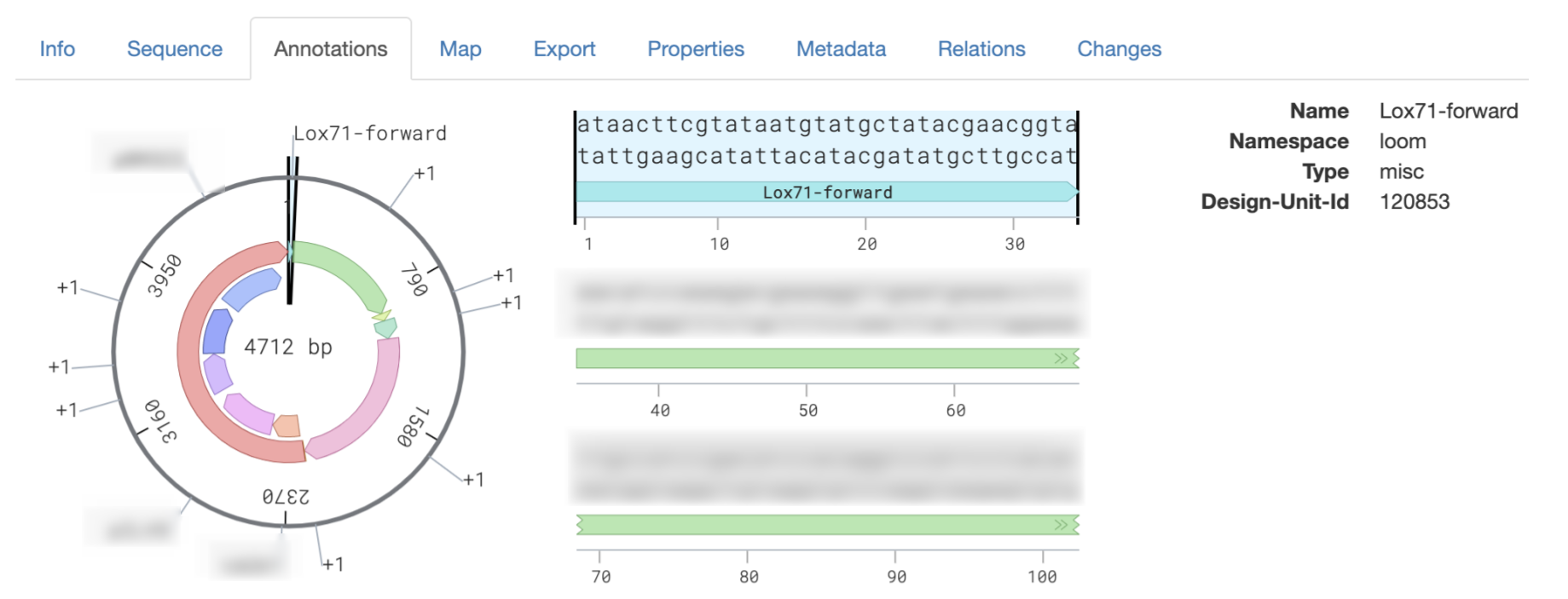
End Result
The aim of my project was to increase the efficiency and efficacy of the cre-lox workflow for biologists at Ginkgo. I produced a new content update step to keep track not only of the changes to molecules in LIMS undergoing a cre-lox recombination, but also of their positional annotations and strain lineages. Now, when a cre-lox operation is performed, scientists no longer have to run a Python package themselves and can rely on OrganiCK to perform the cre-lox calculations and update LIMS for them.
My Ginkgo Experience
Throughout my Ginkgo experience, I was able to develop and foster not only my technical skills, but also my people and whimsy skills. I feel so grateful to have had the opportunity to learn about my team and work on my own project, as well as to have explored all different sides of the company, from product management to scientists in the lab.
I had a great time getting to know the other interns and going to all the fun intern events: from kayaking on the Charles to lunch with the founders of Ginkgo, to going to a Red Sox game at Fenway park! A major shoutout to David Zhou and Hanna Tseng for organizing our events, and a huge thank you to Abe Miller, my Decepticons mentor, for always being there, answering my questions, and cheering me on.
I truly could not have asked for a better internship experience, and have learned an incredible amount this summer. Thank you Ginkgo!
Evan Kenyon, Software Engineering
Introduction
Hi, my name is Evan and I’m currently a rising senior at Duke University, studying Electrical and Computer Engineering as well as Computer Science. This summer, I interned on a scrum team called the “Terminators” working on a project in Loom, Ginkgo’s platform for designing and ordering DNA libraries that is widely used by biologists throughout the company. My work this summer was full-stack, fairly evenly distributed between backend and frontend. My project would not have been as successful as it was without the guidance of my mentor on the Terminators, Travis Beach; contributions from the Product Design team, and feedback from our users.
Main project: Sub-Combinatorial Designs in Loom
In Loom, users design constructs, which are segments of DNA that can eventually be put into an organism for some desired effect. A design in Loom is made up of multiple buckets, and each bucket has one or more design units (reusable DNA parts). Every bucket represents a different type of DNA, so one bucket could be for Ribosome Binding Sites (RBS), another for Open Reading Frames (ORF), etc. Before my project, users in Loom could only create fully combinatorial designs, which means that a construct is created for each possible combination of design units across each bucket.

In the example shown above, 1 * 2 * 2 * 1 = 4 constructs would be created when the user clicks a “Create Constructs” button. The table below shows the constructs that would be created:
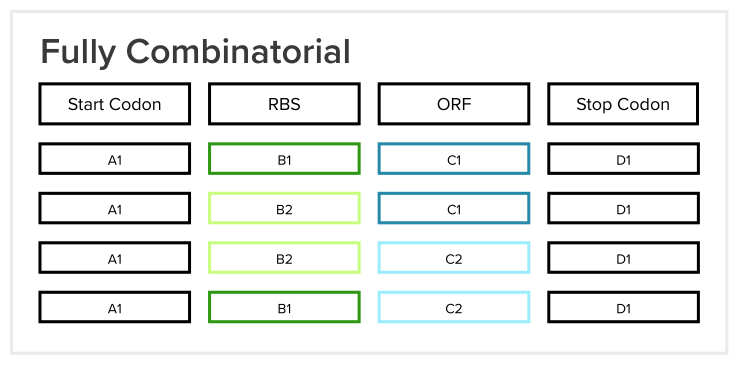
Users were only able to create “sub-combinatorial” designs (designs that included fewer than every possible combination) via our team’s Python library before my project. Since the users are biologists, most of them have little coding experience and have to spend a nontrivial amount of time learning Python in order to access such features rather than simply clicking a button like in Loom.
To make this possible, my project allows the user to upload CSV files defining relationships between design units in different buckets. For example, in the above design, they could decide they only want B1 in the RBS bucket to be used with C1 in the ORF bucket, and for B2 to only be used with C2. Thus, the total number of constructs would now be 2, shown in detail in the table below:

My Ginkgo Experience
Throughout my internship, I had really valuable experiences with fellow interns, my teammates, and other teams alike.
This summer, due to the improved pandemic situation, I was thankfully able to come into the office quite often. This allowed me to meet other people who I would not have otherwise had the chance to meet, and it was great to learn about the many different types of workers at Ginkgo this way. Also, it helped me bond with the other interns since we usually had lunch together at least once a week.
Aside from those in-person office experiences, David Zhou and Hanna Tseng led an intern program this summer that was filled with great social events. This included a luncheon with the founders of Ginkgo, which was my favorite day of the summer. These events also further helped increase the bond among the interns.
Finally, as part of my project, I had the chance to get feedback for my project from the Product Design team during their office hours, and I gained valuable insight into the product design process. I also regularly interacted with my teammates in various meetings and learned about how my team is helping Ginkgo grow.
This summer at Ginkgo has been great, and I have gone through professional and personal growth because of my experience as an intern here.
Stay tuned! In our next blog post, we will be hearing about more cool things that our interns have been doing this summer!
(Feature photo by ian dooley on Unsplash)
