Overview
The potential commercial value of terpenes
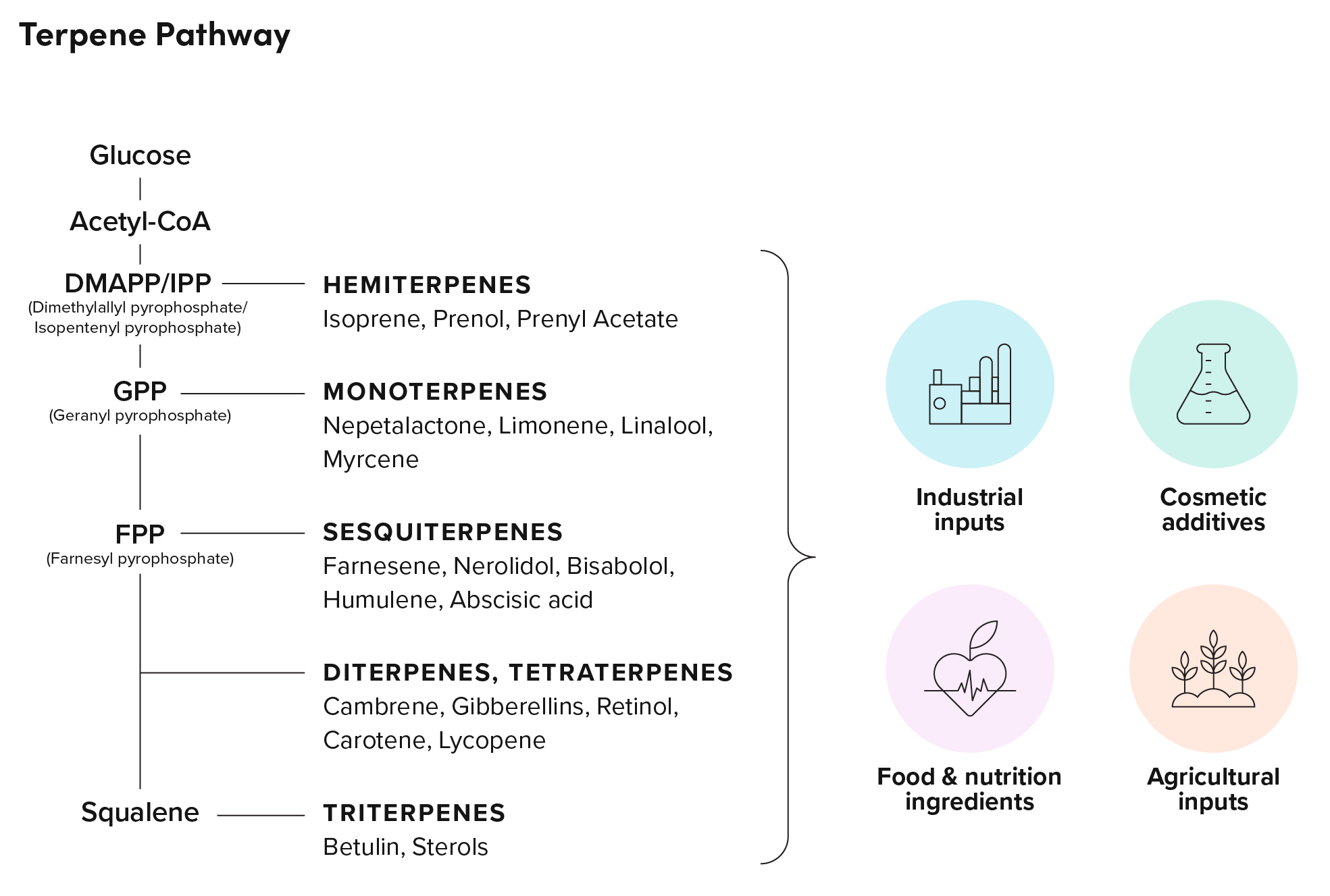
Terpenes, which make up one of the largest classes of natural products, have a wide range of applications across industries. Sesqui-, di- and triterpenes, in particular, have found traction as valued ingredients in the pharmaceutical, agricultural, food and nutrition, and fragrance industries. However, these molecules tend to be produced in low abundance by their native plant or microbial hosts, requiring costly or unsustainable extraction methods that prohibit the full realization of their potential as profitable products.
Synthetic biology research and development has advanced the possibilities of commercially viable terpene production through fermentation. Take the example of alpha-bisabolol, a sesquiterpene found in many plants. With its anti-inflammatory and healing properties, this ingredient has applications in skincare and pharmaceutical products. However, traditional extraction methods from the Brazilian Candeia tree (Eremanthus erythropappus) have yields of less than 0.01%, proving unsustainable with diminishing tree populations (Ribas et al, 2014). Additionally, projected rising market demand will increase the strain on conventional extraction methods, underscoring the need for bioproduction methods. Industry leaders have recognized this opportunity, and through R&D partnerships, have successfully commercialized alpha-bisabolol through fermentation.
Partnering to help de-risk terpene bioproduction
Alpha-bisabolol presents one example of microbial production creating greater market access for a new ingredient. Because biological R&D is inherently risky, we welcome partners to work with us to evaluate the return on investment required to shift production to fermentation. In addition to demonstrating economically viable production at lab scale, partners will need to think through the investment required for fermentation scale-up, product extraction and purification processes.
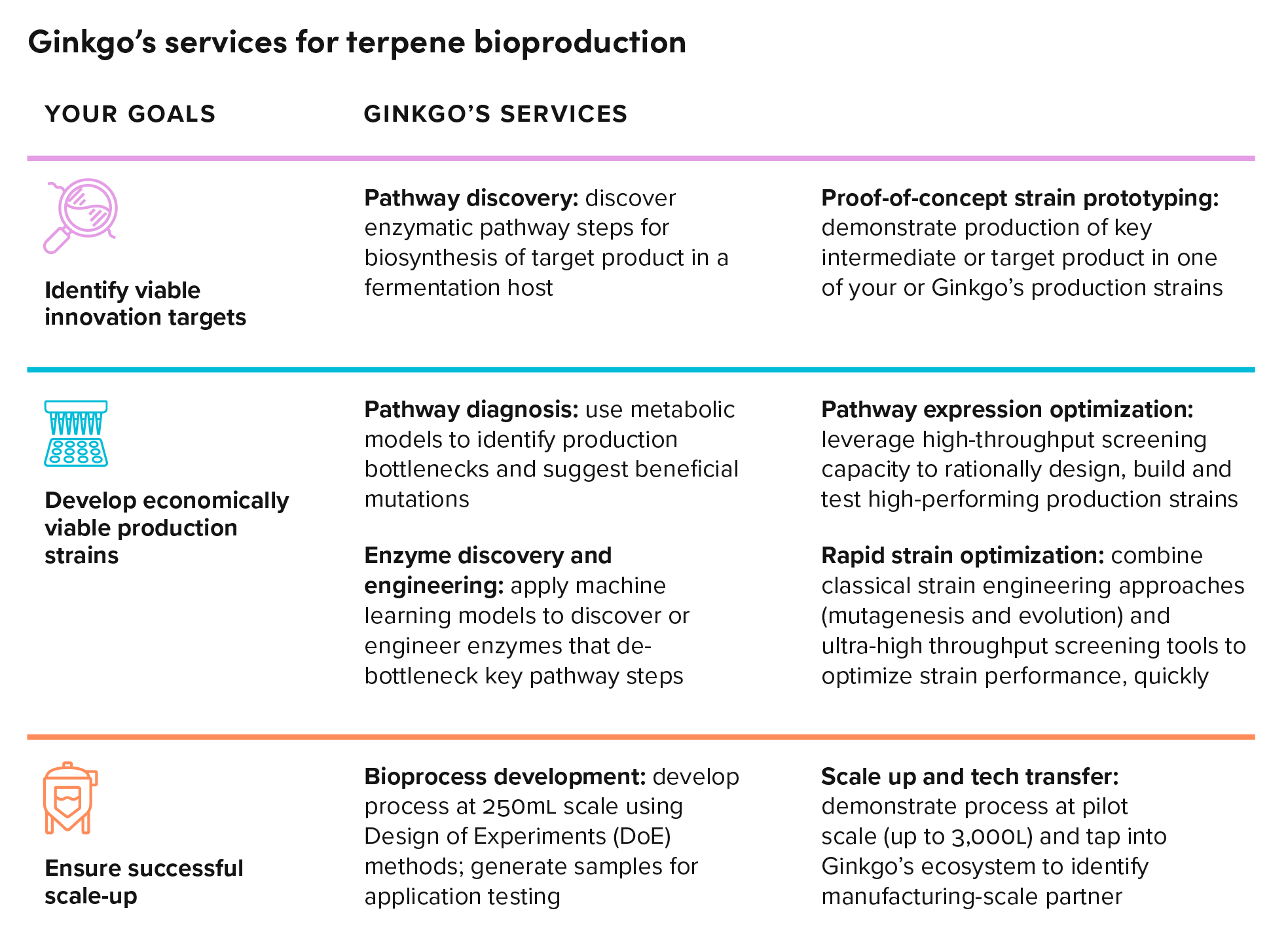
In service of partners looking to develop innovative ingredients, Ginkgo has embarked on several R&D programs towards commercially-viable terpene bioproduction. Through these programs, we’ve gained data insights and honed analytical methods that are readily applicable toward other terpene bioproduction targets. With high-throughput strain engineering capacity, we can readily replicate pathways from plants or fungi into fermentation-friendly microbes and optimize these pathways for commercially viable production. While our commercial deployment team helps prove out an efficient fermentation and product recovery process at pilot scale, we can also identify CDMO partners suited to process scale-up.
Scale and automation power the discovery engine
Outsourcing pathway discovery and optimization to Ginkgo can reduce R&D costs while increasing the chances of achieving target process metrics associated with terpene bioproduction. Through years of partnered R&D work, we’ve developed tools for engineering S. cerevisiae and Y. lipolytica – two “fermentation-friendly” yeasts commonly used for sesqui-, di-, and triterpene bioproduction – in high throughput.
Many high-value terpenes are products of multistep pathways in their native sources. In addition to the enzymatic steps, other genetic elements influence production efficiency, such as accessory proteins and localization sequences. Refactoring complex pathways in yeasts requires significant trial and error. Scale and automation rise to the challenge. Sample large genetic design spaces quickly and efficiently by accessing our highly automated labs.
“Chassis” strains get ahead of toxicity issues
Depending on the end application of the target molecule, partners can access Ginkgo’s suite of proprietary strains, engineered for improved terpene biosynthesis. While terpenes are synthesized from a common pathway in yeast – the mevalonate pathway – each branch comes with its own unique intermediate products, some of which may be toxic to yeast cells or require unique modifications to decorate the core terpene molecule. To manage these issues, flux through the production pathway must be carefully considered and calibrated. Rather than starting this calibration process from scratch with each new program, Ginkgo has engineered a set of strains with high, medium, or low flux through common precursors for sesqui-, di-, and triterpenes.
AI-driven enzyme discovery and engineering overcome roadblocks
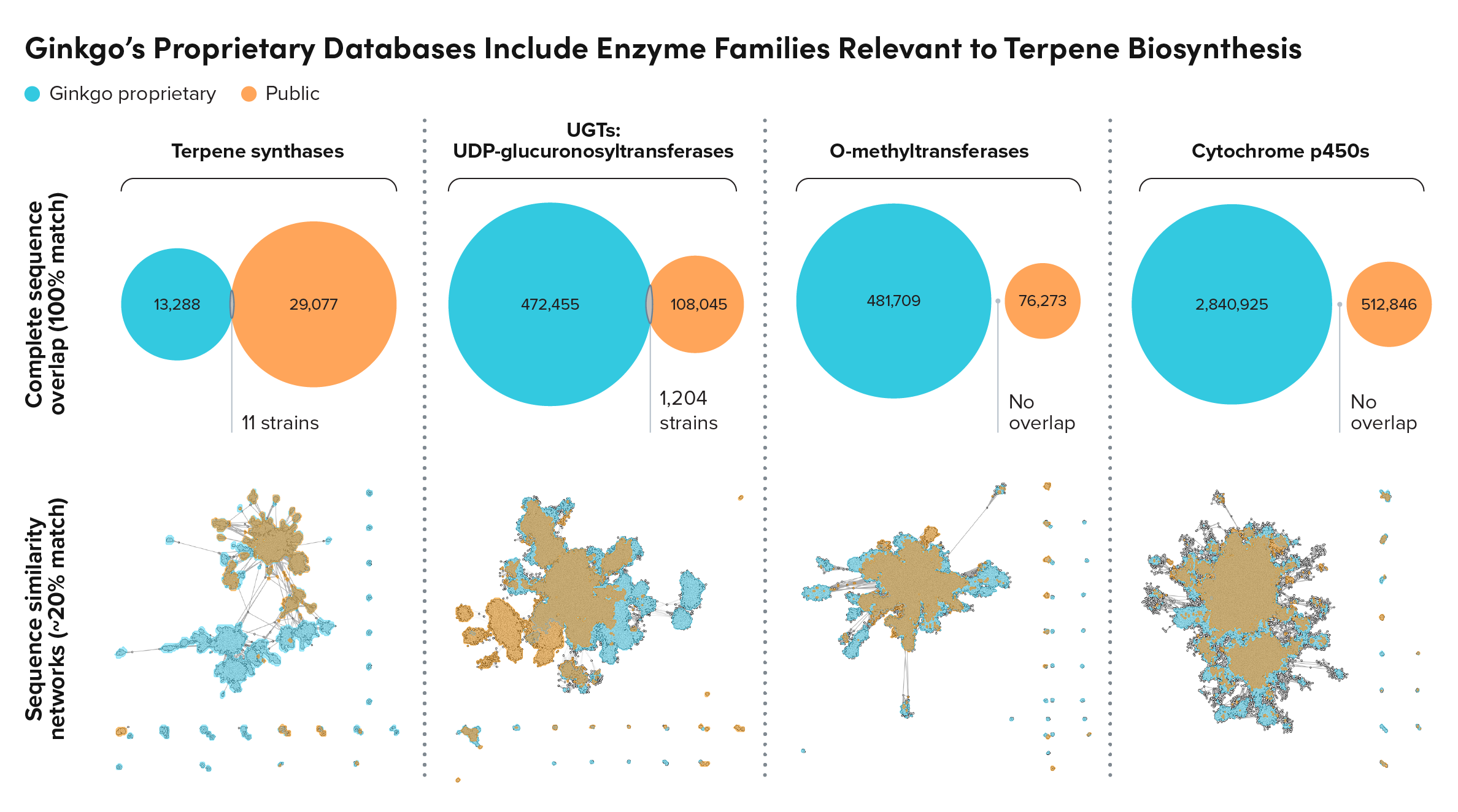
Partners can overcome technical bottlenecks or intellectual property-related hurdles in part through enzyme discovery and engineering. For each enzymatic step along the pathway, improving that enzyme’s rate and / or specificity can unlock step changes in titer. Ginkgo approaches this through machine-learning driven enzyme discovery and engineering. These models use a combination of experimental data from Ginkgo’s >200 R&D programs and metagenomic databases to recommend enzyme designs likely to have improved function.
Along the sesqui-, di-, and triterpene pathways, we’ve done significant metagenomic sourcing and experimentation for key enzyme families, including terpene synthase, UDP-glucuronosyltransferase (UGT) and cytochrome P450 enzymes.
At Ginkgo, our AI-driven design approaches and high throughput screening capabilities allow us to thoroughly search limited or constrained design spaces to achieve program goals. For example, if certain designs or approaches to solving a particular problem are unavailable, expensive, or impractical, we can use our capabilities to design and screen for potential solutions.
Case Study: Pathway discovery to produce a new food ingredient
After sampling a food ingredient extracted from a plant, a partner recognized that this terpene molecule had the potential to establish a lucrative new product line for their company. Academic research had elucidated much of the 9-step pathway beyond squalene that produces this molecule in its native source, but a few steps still needed to be discovered or characterized. This partner approached Ginkgo to serve as their discovery and development engine.
Our team started by screening enzymes from the native plant genome to uncover the remaining steps in the pathway. From this, we identified four pathway steps that involved enzyme families (UGTs and P450s) that have long proven difficult to engineer for better performance. A crucial rate-limiting step involved a P450 enzyme, an enzyme family that has been the focus of much academic and industry research. Through iteratively applying mutagenesis and machine learning models, our team engineered this enzyme. Additionally, our team combed through the native plant genome, and identified several accessory proteins that significantly improved flux through this enzymatic step.
Ginkgo also leveraged its metagenomic library to test several non-native UGTs to improve the rates and specificity for these slow enzymatic steps, as the native enzymes demonstrated low flux and produced many off-target products in yeast. Upon confirming the pathway, our team applied machine learning models to iteratively optimize each enzyme, enabling higher flux towards target product formation.
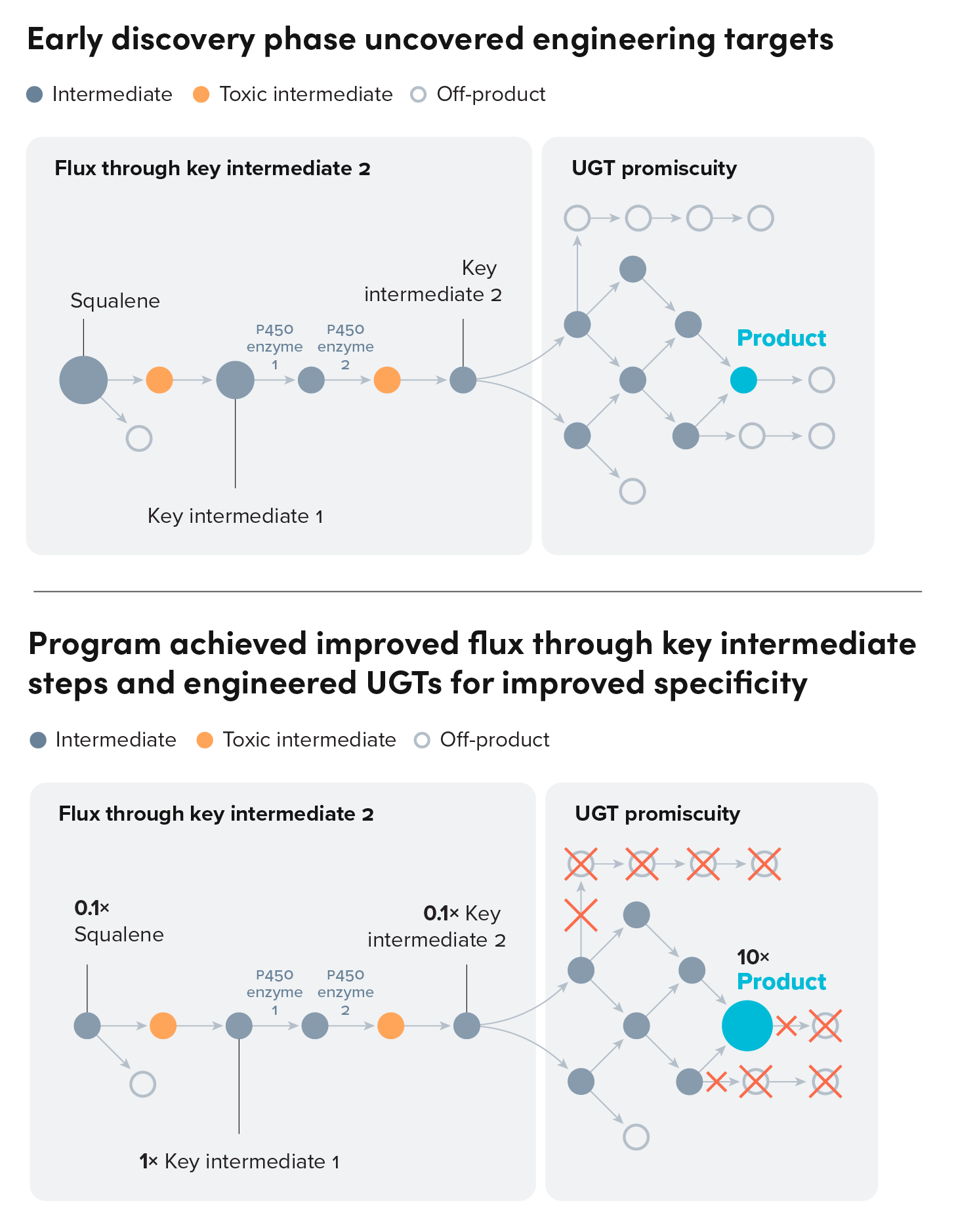
The outcomes of this work addressed the aims of this program and also paved the way for subsequent R&D programs. Several other programs have leveraged variants of the P450 helper proteins and UGTs that were discovered or engineered as part of this program. Furthermore, the data generated by iteratively designing and testing P450 and UGT variants has improved our machine learning models for these challenging families of enzymes.
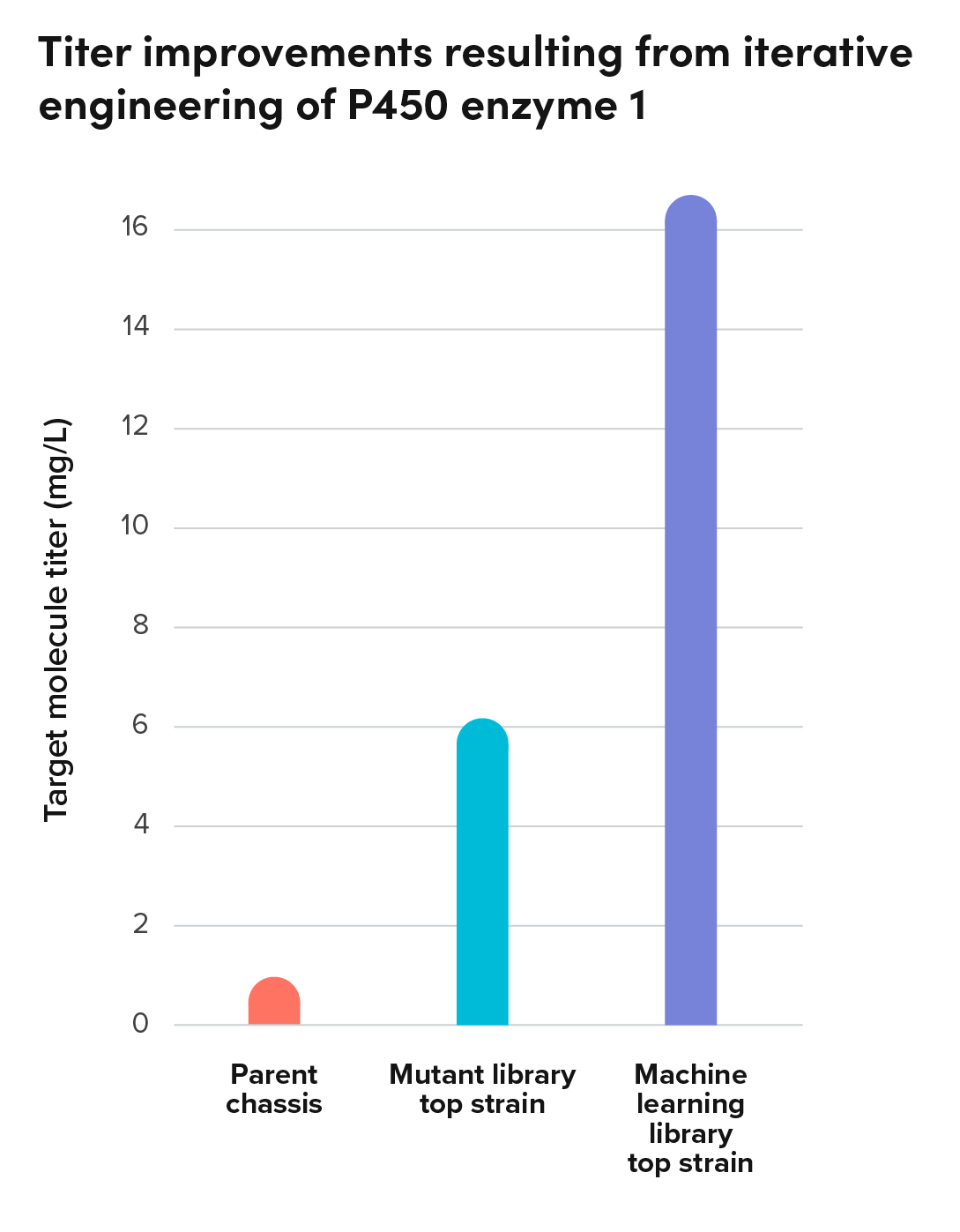
Case Study: Removing a bottleneck through iterative enzyme optimization
A partner commercializing food and flavor ingredients similarly sought to produce a terpene native to plants through fermentation. Scanning the literature, they found that the native enzyme required to convert a precursor to the target molecule also produced a range of off-target molecules. In fact, of all the molecules the enzyme produced, only 13% was the target molecule.
The partner approached Ginkgo to develop a yeast strain that could produce the target molecule with high specificity. Through iterative enzyme and strain engineering efforts guided by sequence and structure-based AI models, we propelled specificity to over 65% of the target molecule, with each round testing between 500 and 1500 enzyme variants for better strain performance.
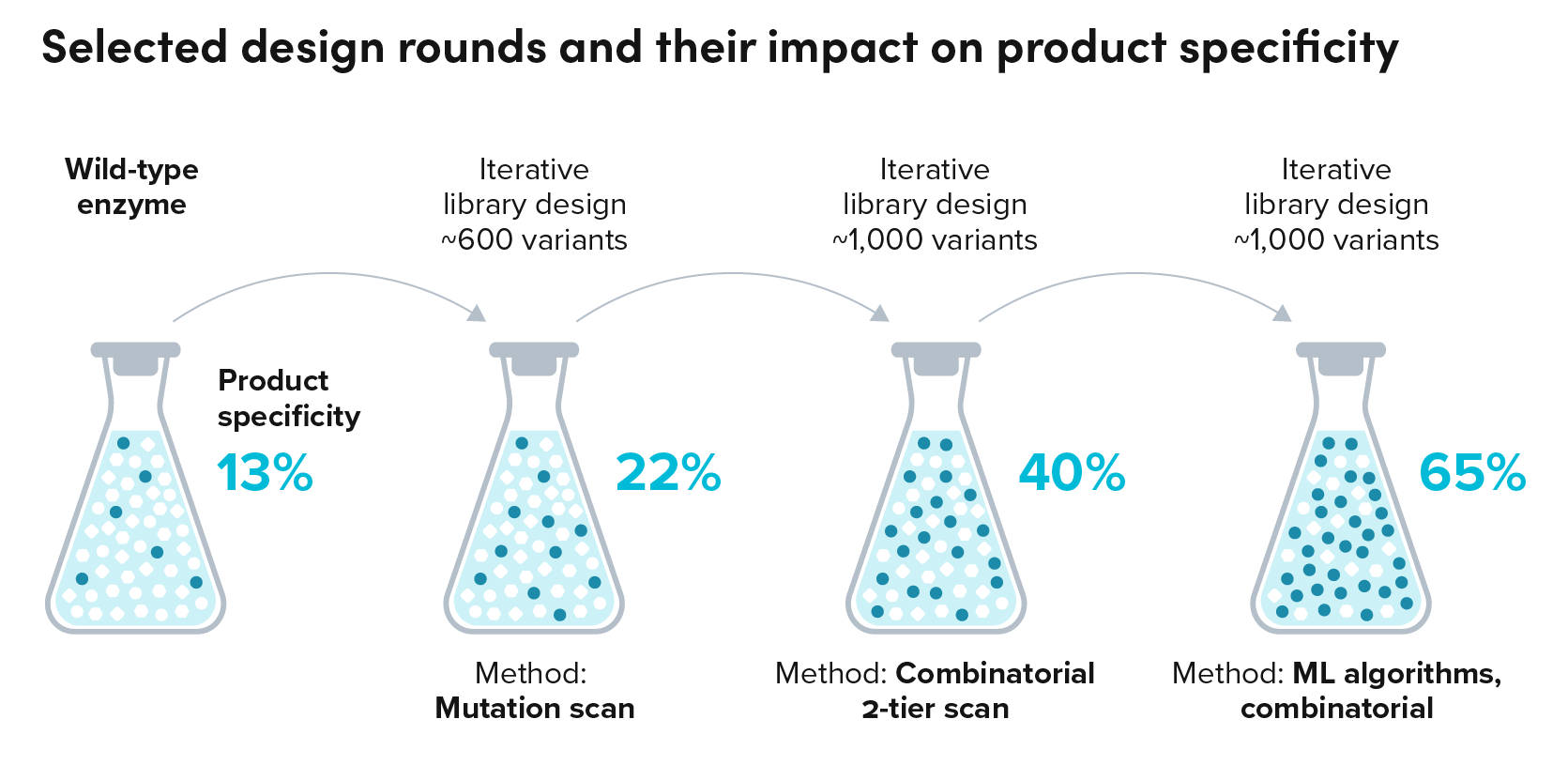
Through this work, we gained quantitative insights into genetic edits that impact flux through the mevalonate pathway. We welcome partners to leverage these data toward their own development programs.
Case Study: Engineering a proof-of-concept strain to produce a pharmaceutical ingredient
In this last case study, a partner approached Ginkgo with the aim of producing a high-value pharmaceutical ingredient, a triterpene natively found in plants. Though another institution had already patented a yeast strain engineered to express the 20-step plant pathway, our partner sought to develop a highly optimized production strain to enable profitable bioproduction of this ingredient.
Ginkgo started with a proof-of-concept program to develop a yeast strain with higher flux through the pathway to 2,3-oxidosqualene. We leveraged outcomes from previous programs – expressing the first 7 of the 20 steps of the full pathway in three different chassis strains, each with varying flux through to 2,3 oxidosqualene. In 8 months, we identified the most efficient strain for demonstrating titers of a key intermediate product.
This program also benefited from the discovery and characterization of cytochrome P450 accessory proteins, described in the first case study. By incorporating these proteins, we could increase the titer of the target without any additional protein engineering. Now that we have demonstrated production of the key intermediate, our next R&D program with this partner will leverage machine learning models and our metagenomic libraries of enzymes to discover and optimize the full pathway toward economically attractive titers.
"*" indicates required fields