Overview
What are the advantages of large metagenomic enzyme libraries? And the dangers of small ones?
Developing large metagenomic libraries allows us to identify top enzyme candidates for functionality as well as variants unencumbered by IP constraints. Use Ginkgo’s enzyme services to search genetic design space to find the right enzyme for your application.
In Summary
- 70
- Hits identified
- 40k
- Genes computationally identified
- 3k
- Genes tested
- 3
- Rounds of Design-Build-Test-Learn
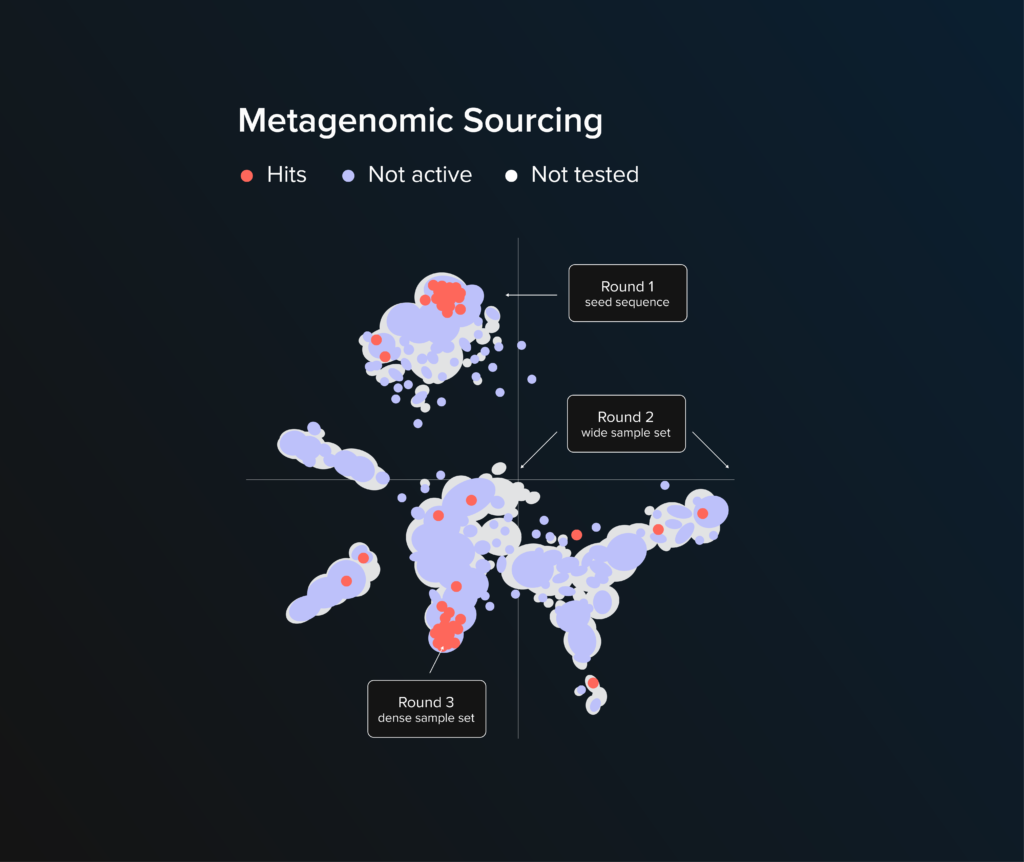
Project Snapshot
Our customer required an enzyme to relieve a bottleneck in a metabolic pathway for a small molecule production host. We identified 40,000 candidate genes in silico encoded for the desired function using proprietary machine-learning metagenomic algorithms. Of these, we tested 3,000 candidates. After three rounds of design and screening, we were able to identify 70 candidate enzymes as hits to meet the customer’s requirements. The diversity of hits expanded our customer’s IP space in engineering this metabolic pathway.
