RAPID OPTIMIZATION OF YOUR PRODUCTION STRAINS
Improve strain performance up to 5x in a matter of months
Techniques like mutagenesis and combinatorial library design rapidly generate millions of strain variants from a starting strain. But conventional methods to screen massive strain libraries are time-consuming and labor-intensive.
Ginkgo’s proprietary encapsulation and screening capabilities (EncapS) can remove this bottleneck thanks to its ultra-high throughput capacity. This service can analyzes millions of strain variants in a single run, increasing the probability of finding a strain with significantly improved activity in a timeline that meets your commercial needs.
TECHNOLOGY DEEP DIVE
Learn more about EncapS: our ultra-high throughput screening technology
Compatibility with a range of hosts
Our EncapS technology can be applied to bacterial, yeast, and fungal hosts as well as mammalian cell lines. We can work with your S. cerevisiae, Y. lipolytica, C. glutamicum, Bacillus, Pichia, and Aspergillus strains among others.
Assay versatility
Screen for a variety of secretion-based properties including improved titer, activity, and yield or decreased product degradation under conditions that are relevant to your application.
Expertise with a range of library generation techniques
Our team is experienced in a wide range of methods to generate strain variants, including classical mutagenesis-based methods. We’ll match the right technique to your needs.
CASE STUDY
Improving amino acid yield through classical strain improvement
Challenge
After multiple rounds of rational engineering, our partner’s amino acid production strain hit a plateau in product yield. They needed to reduce their product costs but had exhausted all known rational engineering targets. We combined random mutagenesis and EncapS to identify a strain with higher product yield.
Outcome
We generated and screened a library of hundreds of thousands of strain variants to identify a new production strain. The partner now uses a top-performing strain from this library that demonstrates a 3% increase in product yield. Because their production strain is employed in industrial-scale fermentation, this yield increase enabled a significant reduction in cost for our partner.
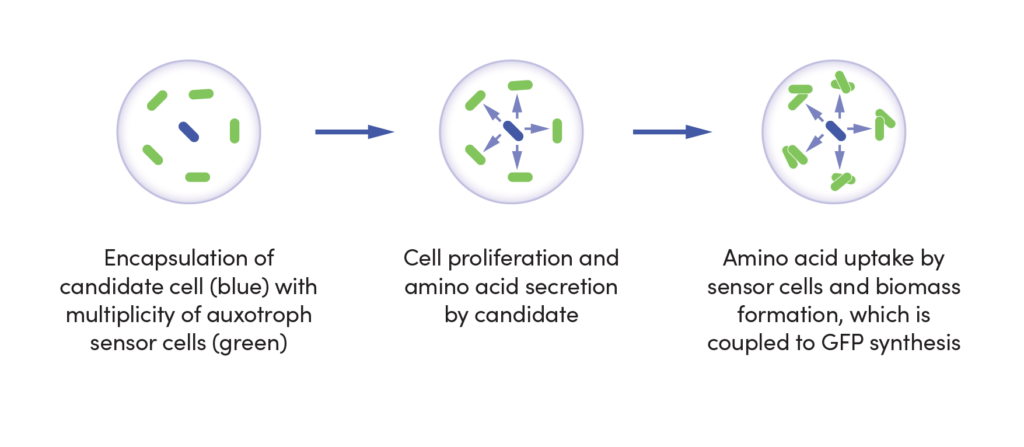
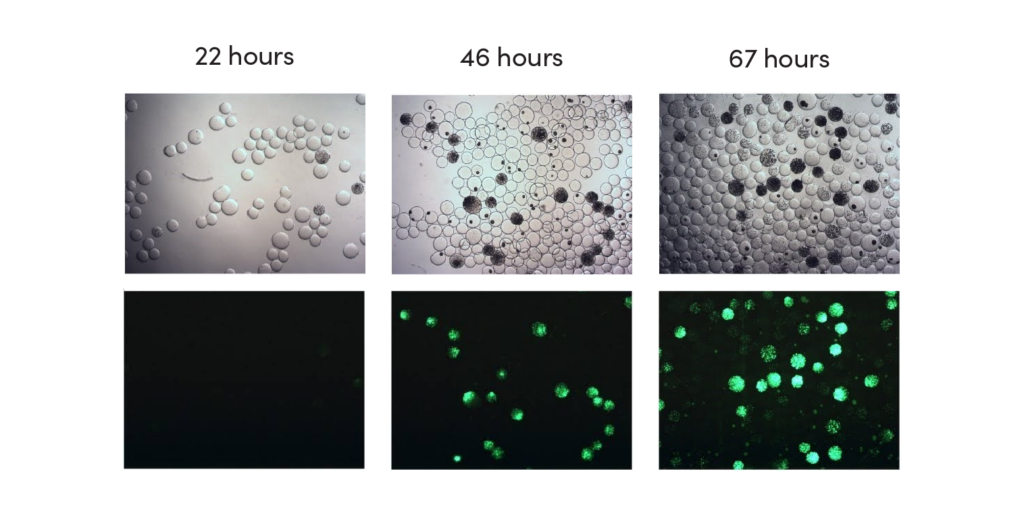
CASE STUDY
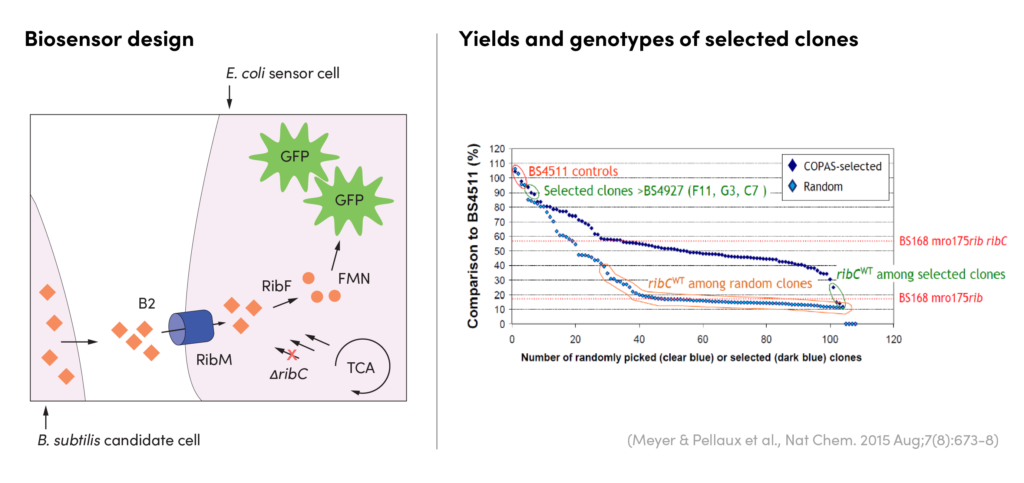
Screening for increased vitamin B2 production
Challenge
Engineered strains employed for industrial-scale vitamin B2 production tend to acquire mutations that impede strain fitness and performance. Identifying these deleterious mutations using traditional sequencing and screening methods would be limited by the throughput of traditional sequencing and screening methods, on the order of only 10,000 variants per week.
Outcome
Our team combined genome shuffling techniques with ultra-high throughput screening to differentiate the mutations that correlate with high vitamin B2 yield from the mutations that impede strain fitness. Specifically, we generated a library of strain variants by shuffling the genome of a production strain with wild type strains. These variants were encapsulated and screened using a biosensor-based assay and EncapS. High performing strains were isolated for sequencing and analysis.
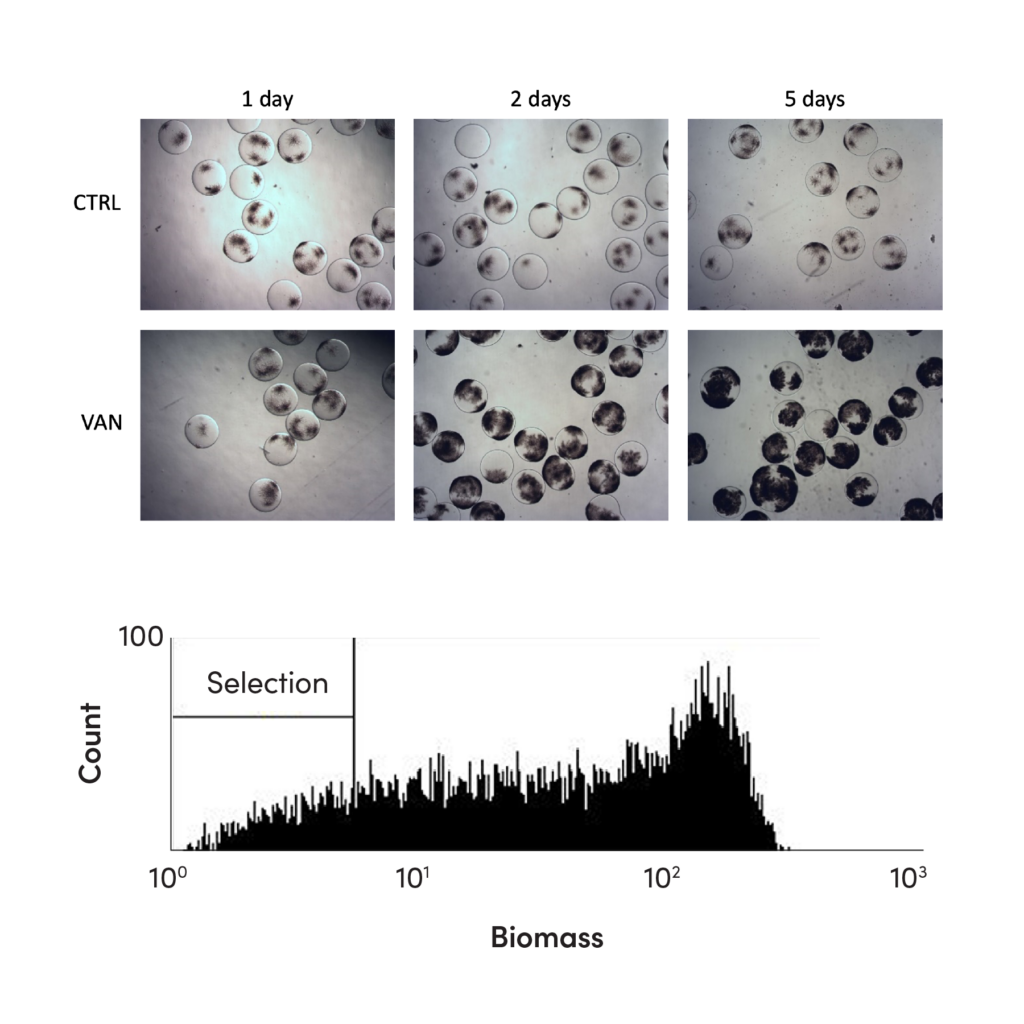
CASE STUDY
Decreasing product degradation of a natural flavor
Challenge
Our partner runs an industrial biotransformation process to produce a natural flavor. The process suffered from limited yield due to product degradation and metabolism within the strain. While they knew which degradation genes they needed to knock out, rational strain engineering was not an option if they wanted to fulfill regulatory requirements for non-genetically modified products.
Outcome
We generated a large library of strain variants using UV mutagenesis. Our team designed a product degradation assay in which strain variants were incubated in nanoliter reactors and fed with media that did and did not contain the flavor molecule as a carbon source. By screening and isolating strains with the lowest biomass formation when fed with the natural flavor, we identified strains with approximately 15% improved product yield and titer at industrial scale.
CASE STUDY
Identification of novel antimicrobial agents
Challenge
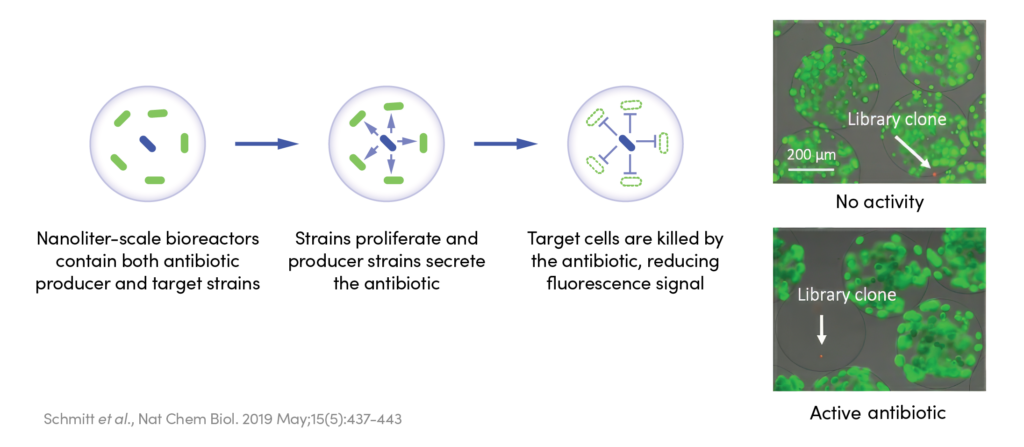
Pathogenic resistance to antimicrobial compounds has been a persistent threat to human and animal health. Our team sought to identify novel lantipeptides with potent antimicrobial activity. This discovery process requires the analysis of large combinatorial libraries to select the best-performing candidates.
Outcome
Our team developed a co-encapsulation assay that combined peptide-secreting host cells with a pathogen model to measure each peptide variant’s killing activity. After running this assay on a large library of peptides, the team identified 126 hits.
"*" indicates required fields