In our last blog post, Roshida and Sean told us about their summer internships with the Digital Tech Team at Ginkgo. This week, Himanshu, Arijit, and Hunter tell us about their internship projects!
Himanshu Reddy, Software Engineering Intern
Hey! My name is Himanshu, and I had the privilege of interning with the Lab Data Processing team this summer. I’m currently a rising senior studying Computer Science at the University of Texas, with a focus on applications to biology. I believe that there is a lot of potential to apply computing to biology, spanning applications from lab automation to AI-driven DNA design. What excites me about Ginkgo is their effort to apply robust engineering practices to biology; it’s a hard challenge, but one worth solving!
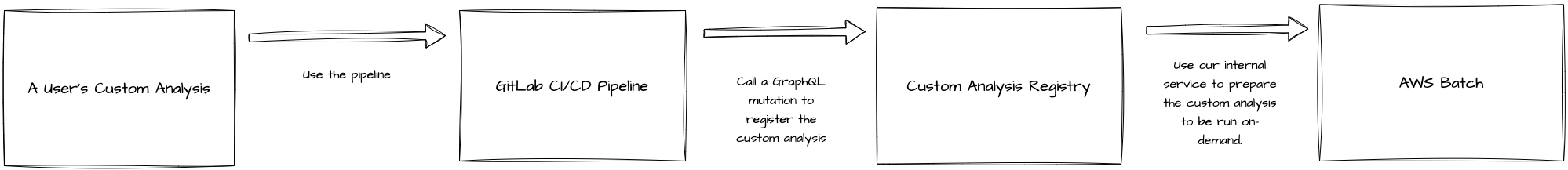
During this internship, I built a registry for Ginkgo’s custom analyses. Custom analyses are scripts written in Python and they allow us at Ginkgo to run custom processing logic on raw bioinformatics data (e.g. counting the number of barcodes in the data from a NGS sequencing run). Existing custom analyses can be made available in Servicely, our portal for foundry services.
However, we didn’t keep track of what custom analyses existed at any given time and what revisions were made to them afterwards. This led to several problems, such as a lack of backwards compatibility and a tedious update procedure when adding or modifying a custom analysis in Servicely. I joined Ginkgo during an ongoing effort to fix many things affecting the custom analysis infrastructure and Servicely. My project, termed the Custom Analysis Registry (CAR), was a part of this larger effort.
I created a Django app that, essentially, keeps track of registered custom analyses. The registration process was handled using a Gitlab CI/CD pipeline in the CAR that could be imported and used in a custom analysis. Once that was done, the pipeline would make a request to register the custom analysis to the registry. The CAR would also do some prep work to set things up in AWS Batch, allowing us to run custom analyses on-demand. This allowed us to keep track of the custom analyses at Ginkgo and make the custom analysis development process much better!
My internship was a very enjoyable experience! I was lucky to work with smart, motivated, and supportive people who helped and advised me during my internship. The people team also hosted many great events for the interns, like the tour of the DNA synthesis facilities in Ginkgo’s Cambridge office.
One thing I liked about Ginkgo as a whole was their open and collaborative culture. Because of it, I was able to attend many of the office hours sessions hosted by different teams. I was also excited to learn about the deep learning initiatives happening at Ginkgo, as deep learning in biology has really taken off in the past few years. I’m excited to see how biotech companies like Ginkgo incorporate AI into their workflows.
Overall, my internship was a great experience and I’d highly recommend Ginkgo Bioworks as a place to work!
Arijit Nukala, Software Engineering Intern
Hello! My name is Arijit, and I’m a rising senior at Johns Hopkins studying Biomedical Engineering and Computer Science. This summer I worked on the Operator Guide team, which owns Ginkgo’s in-house user interface for managing laboratory tasks. My project involved building a “Pause State” for Ginkgo’s lab operators.
In the lab, operators execute on the workflows that underlie specific Ginkgo Foundry services. Each workflow involves a different instrument and process. Understanding how long each of these processes takes is essential to efficient planning of Ginkgo’s projects. The goal of my project was to support the ability for an Operator to pause work after it has been started in order to more accurately calculate the turnaround time (TAT) for Ginkgo’s services. Operators needed a way to pause this clock so that requests that are blocked because of things outside the Foundry’s control don’t artificially inflate the turnaround time for the service.
For my project, I developed a Pause Annotation model and API for manipulation of data in Django, a common and powerful Python web framework that integrates with front end systems such as Ginkgo’s React based Portal-UI. The data model I created links to workflows being processed by Operators and tracks the pause time and pause reason each time work is paused. When a workflow is resumed, the resume time is also logged. Because each pause/resume for a workflow is stored in the data model (similarly to lap times on a stopwatch), we can calculate the total pause duration and working duration. All of the working or unpaused times can be added together to get the TAT for a workflow.
Originally, I planned to implement a more general “annotation” feature. Each annotation corresponded to either a “pause” or a “resume” action , and was generalizable for annotation features beyond pause/resume in the future. However, calculating TAT from data structured as two entries for each pause/resume cycle is complicated and computationally intensive. I instead opted for one entry that contains the pause and resume datetime to simplify TAT calculations later.
At the end of my internship, I was able to release my feature company wide and received positive feedback on it. I added additional features as part of an MVP+ release, presented my work to Ginkgo’s foundry teams, and created a Tableau dashboard to visualize pauses in batches. My feature had a greater impact than I could’ve imagined was impactful despite my short stint as a developer.
Being part of the Operator Guide team this summer was an amazing experience. I had a chance to see how software development teams ideate, organize, and execute their work. During my daily work and through feedback, I learned about intelligent software design, system efficiency, and the amount of teamwork that goes into designing great software.
I’m grateful for the opportunity to stay in Boston this summer and work in Ginkgo’s incredible Seaport-based office. Besides consuming enough cold brew and blueberry cobbler protein bars to last me a lifetime, I had a chance to meet other like-minded interns, develop friendships with Ginkgo employees, and enjoy Ginkgo’s fun-spirited work culture. Making biology easier to engineer through software while having a little fun along the way was a dream come true for a biomedical engineer and computer scientist like me.
Hunter DeMeyer, DevOps Intern
Introduction
Hi! My name is Hunter. I’m a graduate student studying Bioinformatics at the University of Illinois. This summer I joined the Cloud Infrastructure team to gain experience in operations so that I will be better equipped to build end-to-end solutions for scientific problems. I worked on two projects while at Ginkgo: a data visualization and business intelligence tool for surplus cloud spending and a Large Language Model for classifying DNA sequences.
AWS Cost Reduction
First, AWS cost reduction. At Ginkgo, we give our science-oriented teams a lot of freedom when it comes to provisioning the resources they need to get their work done. This lets each team independently build infrastructure as required which is great for keeping projects on schedule; however, sometimes the resources chosen are overkill for the task at hand or the resources are not shut down when they are no longer needed. Complicating things further, we follow AWS’s recommendation of using sub-accounts to isolate different workloads and enhance security. With hundreds of AWS accounts serving different teams, wasted dollars were extremely hard to track down across our entire organization.
Luckily, AWS provides a partial solution for this problem in the way of Trusted Advisor Reports. Each report focuses on a specific service and a specific optimization for it, such as cost or security. Our top interests were in Low-utilization EC2 Instances, Underutilized EBS Volumes, and Idle RDS Instances. These reports are specific to each account.
My project provides a cloud-native method for extracting and visualizing Trusted Advisor Report data. On a regular schedule, an AWS Step Function runs a lambda to log in to each AWS account and pull its Trusted Advisor Reports in parallel. These reports are then paired with some additional account information and certain fields are parsed into the proper format. From there, the step function kicks off an AWS Glue crawler to pull the reports into a Glue database. Finally, an Amazon Quicksight dashboard renders the data into interpretable and interactive visuals.
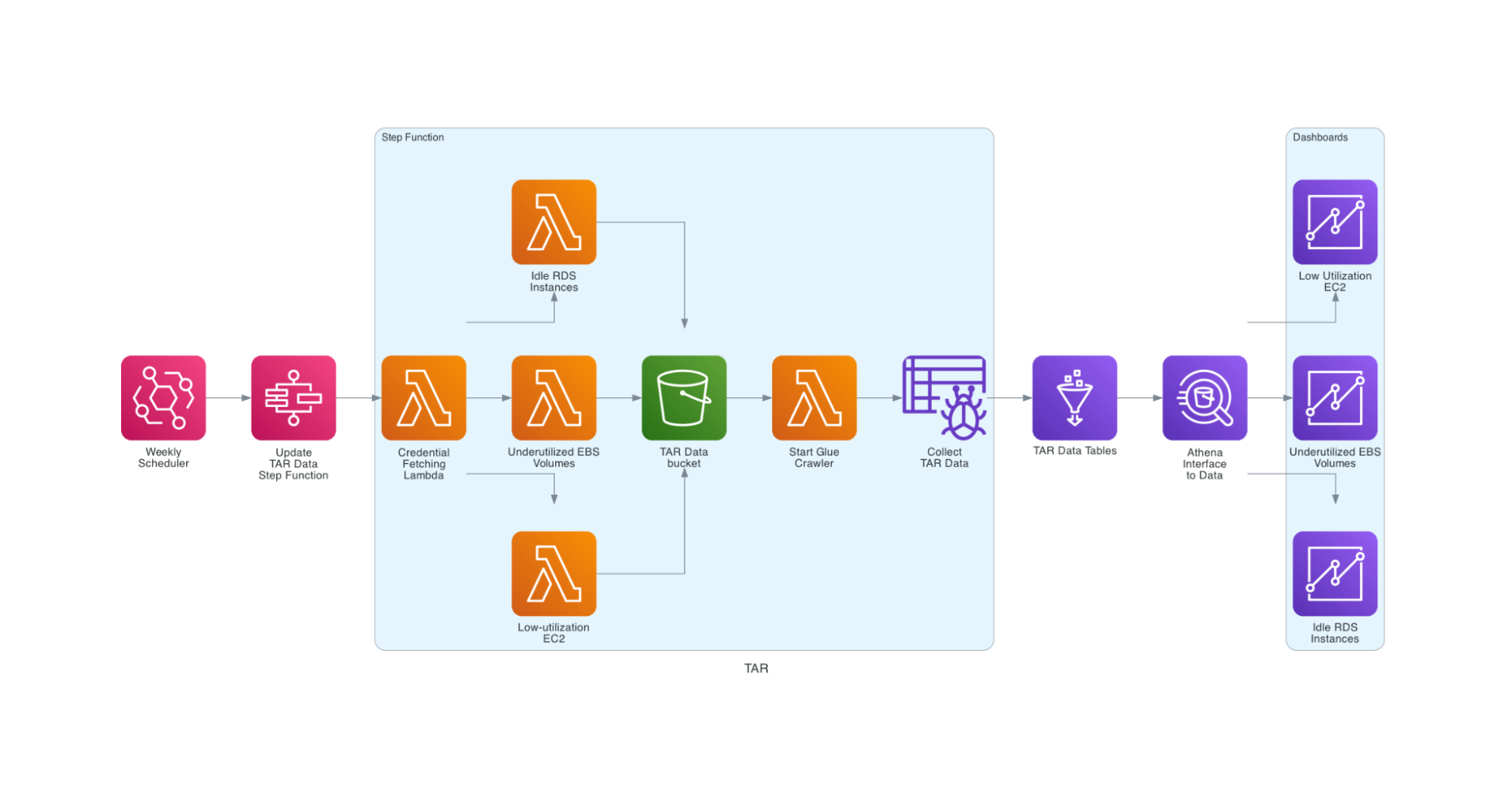
This solution provides Digital Operations management with weekly insights into resources where spending could be optimized, so they can quickly be shut down. On the Cloud Infra team we believe that nothing should be deployed manually, so we try to automate everything as much as possible and in this case it means creating Infrastructure as Code (IaC). The entire stack of lambdas, schedulers, step functions, databases, and analyses can be recreated within minutes from an AWS Cloudformation template.
This internally developed observability platform is exciting in its own right, but it can be expanded to offer so much more in the future. In addition to visualizing data, it could also leverage the existing event-driven infrastructure to cull over-provisioned resources without human intervention, automatically keeping our cloud infrastructure as lean as possible. I’ve also written some code to downsize or terminate EC2 and RDS instances. Right now it needs to be manually triggered but eventually we’d like to have an event-driven process that can automatically reduce costs using this method. Maybe a future intern project!
While I found my Cloud Infra role rewarding, it was not as close to the science as I would want to be in a long term role. My manager Dima Kovalenko was kind enough to let me explore some of the scientific work going on at Ginkgo through a collaboration with our Vice President of AI Enablement Dmitriy Ryaboy.
AI-powered DNA Sequence Buildability Classification
One core tool we use at Ginkgo to engineer biology is DNA synthesis. Sometimes this process is straightforward and DNA fragments are easy to produce, but other times synthesis isn’t yet possible and will not be attempted. We use certain heuristics to analyze every piece of DNA requested for synthesis, and determine whether it will be buildable.This is a great example of a binary classification task, and is one that we have a ton of good-quality data for. Moreover, we have data on why sequences get rejected, as well as the relatively rare DNA sequences that our checks pass but fail to synthesize correctly. This dataset is a good candidate for experimenting with Large Language Models for biology, and comparing them to current processes in use at Ginkgo.
Large Language models (LLMs) work by “tokenizing” inputs; grouping frequent sets of symbols together and counting the number of occurrences of each one. Generalized LLMs (think ChatGPT) have a high number of symbols, and operate on relatively short sequences of text. Biology is different though; the diversity of a sequence is much lower with an alphabet cardinality of only four. Additionally, biological strings can extend to be millions of bases long. Until recently, these requirements exceeded the capabilities of LLMs. We need a specialized model that is both sensitive to individual bases and able to handle extremely large contexts.
HyenaDNA is a recently proposed LLM for biological string analysis. It replaces the usual transformer architecture of LLMs with “hyena” layers. Hyena forgoes the traditional attention mechanism, replacing it with a subquadratic convolution-based technique. This enables much longer sequences or much larger token sets. HyenaDNA takes advantage of these layers to dramatically increase the maximum possible length of an input sequence all the way to one million characters. We decided to experiment with HyenaDNA and see how well the pre-trained model performs on the buildability classification task, using both fine-tuning and in-context learning.
We have a huge amount of historical buildability information; the most difficult part of this task was extracting those data from our databases and formatting in a way that made it possible to ingest into the model; DuckDB was super helpful for this. In addition to that, we needed to ensure that the training data was not skewed to favor a particular time frame, organism, or error class. For this I had to compute a joint probability distribution that weighted each sample based on the rarity of its fields. Once the data was prepared, I stood up a powerful EC2 instance and fine tuned the hyenaDNA mode, achieving about ~80% accuracy in preliminary tests. I wish I had more time to work on this, but I’m very happy with the progress I made on it.
Final Thoughts
I had an incredible summer at Ginkgo and learned so much about being a DevOps engineer. I now know that it isn’t what I want to do long term, but am excited to apply the skills I’ve learned in future roles. I’m extremely grateful that I got to explore such a wide breadth of work at Ginkgo. I’m also very appreciative of the hard work put in by the Early Talent team, who organized tons of fun and informative events throughout the summer. Some highlights include a Boston Red Sox game, an Ask-Me-Anything session with CEO Jason Kelly, and a trip down to Cape Cod. Every bioworker I interacted with over the summer was excited about the intern program and more than happy to provide some help or just answer questions. The passion people have here is unparalleled, and I consider myself lucky to have been a part of this company for the past three months.
(Feature photo by Saiph Muhammad on Unsplash)
