Automated ALE harnesses the power of evolution — Part 1
Adaptive Laboratory Evolution (ALE) was developed in the mid-20th century, but it’s only recently that scientists have been able to leverage this process for industrial partners. Ginkgo’s Head of ALE, Simon Trancart, discusses how Ginkgo uses ALE as a fast, unbiased strain development tool that is powerful on its own or paired with a metabolic engineering campaign.
Humans of Ginkgo Bioworks is an interview series featuring Sudeep Agarwala interviewing some of the brilliant folks at Ginkgo to learn more about the technology that makes our work possible.
— This is the first part of a three-part interview.—
Read Part 2, Inside ALE, here
Read Part 3, Applications and Opportunities, here

Sudeep Agarwala: You’re in charge of Adaptive Laboratory Evolution (ALE) at Ginkgo — a way for guiding evolution in the lab. Why is this something that’s important for a company that engineers strains? Why is guiding evolution in the lab an important tool for metabolic engineering?
Simon Trancart: So the beauty of ALE or other “artificial selection techniques” that try to mimic natural selection is that you don’t need a priori knowledge on what is the bottleneck or what mutations will be required to optimize your pathway. So the best fit for ALE is when you have a strain that you want to improve, but you don’t know how.
Or you might know where you would play with genome engineering, but you cannot because there are no tools for an exotic organism that we can’t engineer easily. Or if you need a non-GMO application.
So I would say, that’s what the most obvious applications are: things that are hard to engineer or can’t be engineered. The very important limitation is that it must be related to fitness, growth or survival.
SA: I noticed you mention “other artificial selection techniques” — so ALE is not the only way to exert natural selection in the lab?
ST: There are multiple ways to mimic natural selection at the lab for the purpose of directed evolution of cells or entire genomes. One approach consists in two sequential steps: diversity generation and then screening, that’s what the EncapS team at Ginkgo does–create a library, which is then screened in ultra high-throughput. In ALE, diversity generation and screening both take place during continuous cultivation. ALE takes advantage of genetic drift in a population and allows the variants that arise to be subjected to natural selection through continuous culturing.
Not all ALE methods are the same. Let’s take Richard Lenski’s work as a famous ALE example. Since 1988, Lenski has been conducting what’s known as a serial-passaging experiment, repeatedly transferring E. coli from one container into another container containing fresh media to observe evolution over thousands of generations. His manual approach over three decades has yielded remarkable insights into microbial evolution.
SA: So ALE is a way to capture this? To mimic natural selection in the laboratory?
ST: Yes. But there are other ways to implement laboratory evolution–or adaptive laboratory evolution, ALE. The picture here shows an implementation using continuous cultivation in a single vessel. This type of system was first implemented in the late 1940’s. There was one team led by Aaron Novick and Leo Szilard at the University of Chicago, and another team in France by Jacques Monod at the Institut Pasteur that really understood that we can evolve microbes quite fast if we cultivate them continuously under controlled conditions.
SA: So these fermentation methods can actually evolve a population of cells to do what you’d like?
ST: Well it’s interesting what you’re saying, because you’re talking about fermentation. We do not see our system as a fermentation tool. We could say that fermentation aims at optimizing the output of one genome and you play with the conditions to optimize the output from that genome. Whereas evolution–ALE–aims at producing an optimized genome from a starting strain or library and you will play with the conditions that will direct adaptation to those conditions.
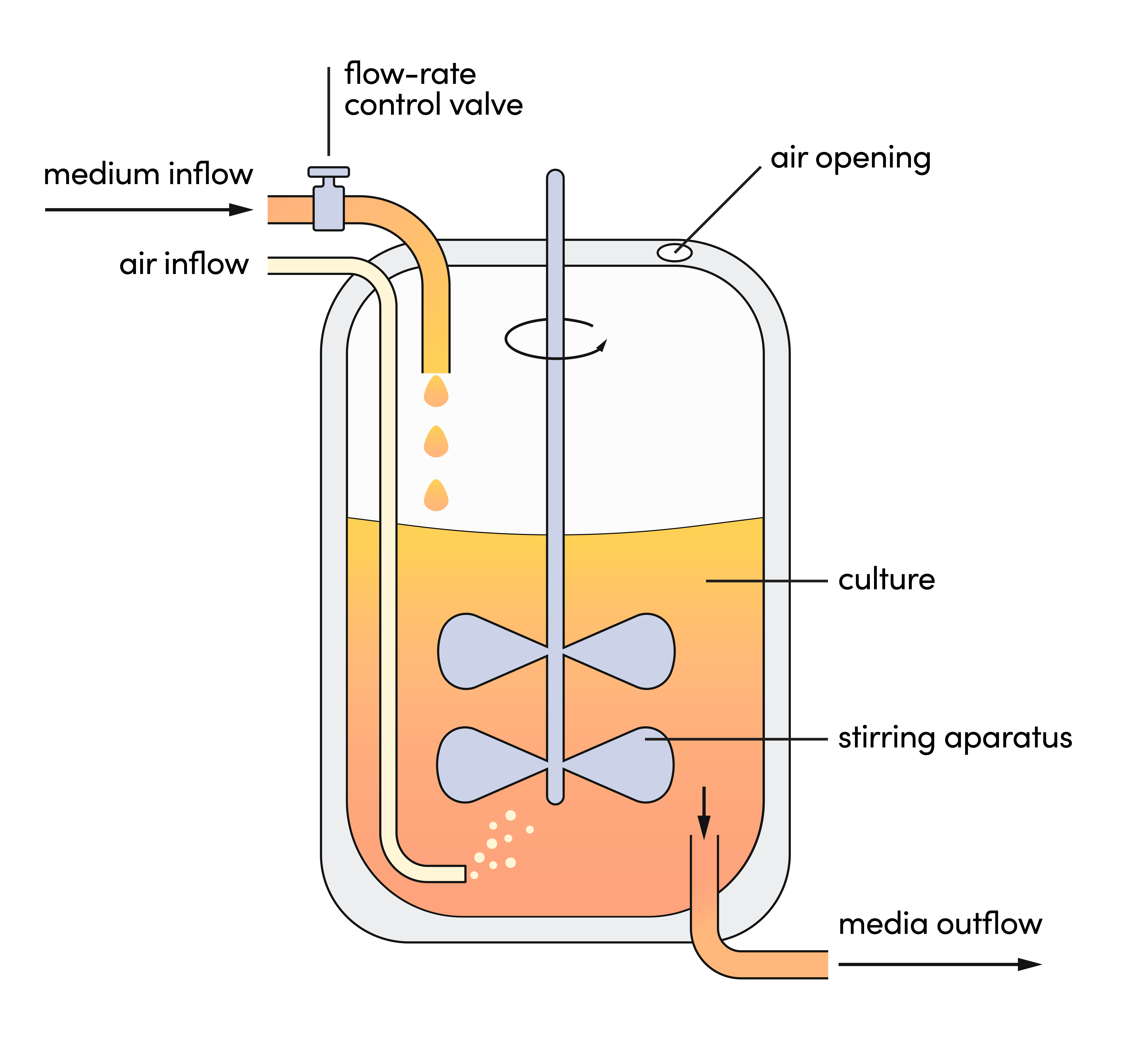
Having said that, though, it’s important to note that you cannot select for whatever trait you want, but for better fitness under specific selection conditions.
The other thing to point out is that people often use fermenters or liquid handling robots in an attempt to automate ALE. And that’s interesting–that people have taken equipment designed for a given purpose and used it to try to make ALE into a system that is automated. But for many reasons like contamination, biofilms or maintenance requirements, this type of method can have drawbacks and issues associated with it. It simply does not work if you want to really automate ALE and that’s the reason why we designed a system specifically for this purpose.
SA: What are some examples for how ALE would work?
ST: For example, if we want to make a strain grow better in a set of given physical and chemical conditions, ALE is a right fit. So: increase the growth rate on the given medium, change media, adapt to new media, new carbon sources, new nitrogen sources, adapt to toxic chemicals, increase tolerance to toxic chemicals, to extreme pH conditions, adapt to higher oxygen tolerance.
These are very basic applications. And I do see a lot of synergies with actually rational engineering. Because any time that you would modify the genome of a strain it will, most of the time, be at the expense of some fitness, especially if you’re modifying a lot of genes. But you can recover fitness after genetic modifications using ALE to stabilize the genome for further engineering.
SA: So ALE is a tool that can be used right alongside more conventional strain engineering?
ST: One of the most beautiful examples is when you can actually engineer a new synthetic activity that will be coupled to growth. So ALE is a great tool when your engineering is substrate-related. So, for example, “I would like to clone heterologous enzymes to utilize C5 sugars, for example, and improve that using ALE.”
There might be other opportunities where rational engineering methods are used to couple the targeted activity to growth. And then: use ALE as a lever to fasten the implementation of synthetic activities into the organisms.
SA: We’ve spoken about working with different organisms in ALE. What organisms have you worked with in this system?
ST: We have run many projects with different bacteria, different yeasts, and a few microalgae.
We had one project with plant cells where it worked well. It was maybe not a good fit because the doubling time was like four days or so. So you can imagine that evolution takes more time, right? But at least we could demonstrate that we can continuously cultivate this kind of cell during–I think it was three months. It was gratifying because it proves that our system really does not contaminate. After so many hours, any contaminant will dominate here. We did that on the rich medium. And we didn’t see anything.
SA: Any highlights?
ST: We had a collaborative project on Pseudomonas putida as part of a collaborative project funded by the European Commission. We were invited to join this by another group that had developed a bacterial chassis aiming at producing bio fluoro polymers.
And they had designed a way to both produce these polymers in P. putida, as well as to couple the fluorination to growth. So, to be clear, they had a scheme where the bacteria cannot grow if it doesn’t incorporate fluorine in its metabolism. And then the fluorine would be directed towards production of fluoro polymers. That was a beautiful synbio project where we could demonstrate the power of combining rational design with ALE to implement new-to-nature activity in life. We tackled other problems with ALE, notably because P. putida doesn’t naturally grow on high levels of fluorine.
And so we did several ALE campaigns in that program, some for improving tolerance to fluorine/fluorinated compounds, which are highly toxic, and others, which aimed actually at improving the growth of strains that were dependent upon the uptake of a fluorinated compound.
It worked pretty well. And we believe this is the type of approach that could be developed for other applications.
Read Part 2, Inside ALE, here
Read Part 3, Applications and Opportunities, here
 Simon Trancart joined Ginkgo through the acquisition of Altar, a French biotech company he co-founded and led as CEO. Altar specialized in automated adaptive laboratory evolution (ALE), a niche that Simon navigated with his background in engineering and civil engineering.
Simon Trancart joined Ginkgo through the acquisition of Altar, a French biotech company he co-founded and led as CEO. Altar specialized in automated adaptive laboratory evolution (ALE), a niche that Simon navigated with his background in engineering and civil engineering.
At Ginkgo, Simon leads the Adaptive Laboratory Evolution, based in Évry-Courcouronnes, France. Simon’s work focuses on the automated ALE process, which the performance of ALE campaigns. He has been instrumental in integrating the ALE team’s work with Ginkgo’s foundry services, enabling better execution and insight into ALE. Simon’s expertise extends to the application of ALE in various organisms and its coupling with rational design.
