Automated ALE harnesses the power of evolution — Part 2
Ginkgo’s head of ALE, Simon Trancart, discusses how ALE at Ginkgo with the Genemat technology overcomes the challenges of contamination and biofilm production. Ginkgo leverages ALE to deliver genetic variants that have been carefully shaped by natural selection in the laboratory.
Humans of Ginkgo Bioworks is an interview series featuring Sudeep Agarwala interviewing some of the brilliant folks at Ginkgo to learn more about the technology that makes our work possible.
— This is the second part of a three-part interview.—
Read Part 1, Why ALE?, here
Read Part 3, Applications and Opportunities, here

Sudeep Agarwala: Adaptive Laboratory Evolution — ALE — is a powerful tool that’s been around for a long time — I believe you said the 1940’s. Maintaining a culture under a constant growth rate for weeks, months, even years, harnesses the power of evolution for strain improvement campaigns. But making an automated system has remained challenging. Why is that?
Simon Trancart: The idea behind laboratory evolution is that you keep a suspension growing permanently.
One way to do ALE is to do serial passaging, which involves the sequential transfer of microorganisms from one growth medium to another. When the culture is transferred to a fresh medium (a passage), only a small portion of the culture is carried over. If a mutation confers a fitness advantage, the organisms with that mutation will grow and reproduce more quickly than those without it. Over time, they will make up a larger and larger proportion of the culture.
Another–I would argue, more effective–way is to cultivate continuously at constant volume. You add sterile medium to sustain growth; you also have to withdraw the same volume that you added. And that creates a competition, because there is growth on the one hand, and you have dilution of the population on the other hand, and so only the microbes that grow at a given base rate will actually have a probability to survive and transmit their genetic heritage over time. Here also, a beneficial mutation will progressively dominate in the population.
When you attempt to automate one or the other way to do ALE, you may face contamination issues. For ALE, you want something to be very, very reliable. And beyond contamination, there are other issues. In continuous culture in a single vessel, you will have biofilms, whereas serial passaging is really hard to automate. We know that robots need maintenance and if you want to explore thousands of generations, you might really be exposed to failure and interruption of your experiment.
SA: But you’ve found a way to reliably automate this?
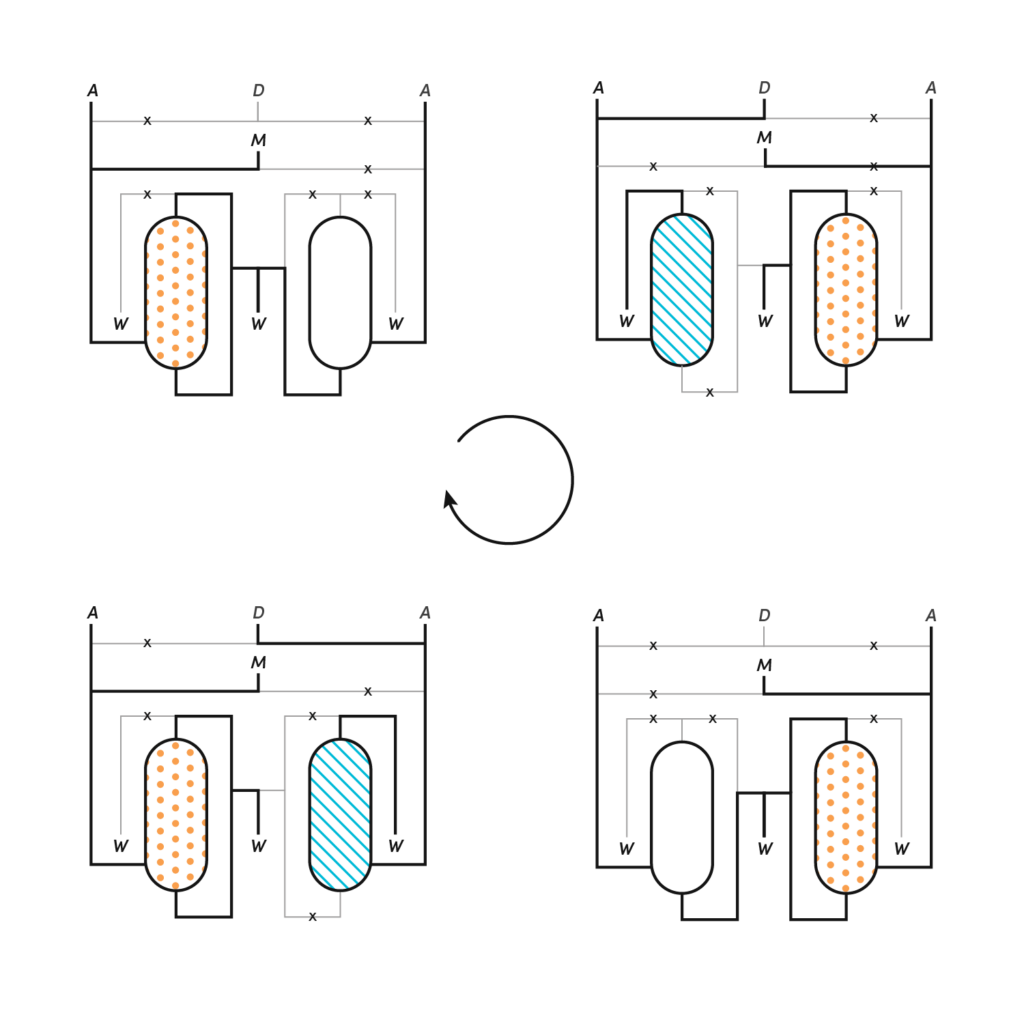
ST: Yes, and this required tackling critical issues: biofilms and contamination. In any system where the culture is being maintained in a single vessel, eventually, you will get a biofilm at some point.
That is, nature finds an easy way to cheat the system. Over time, in order to stay in the vessel and escape selective pressure, cells will stick to the vessel wall. Evolutionarily, it’s very effective–finding a physical way to remain in the population. Everywhere you have this kind of long-term cultivation under selective pressure, you will find biofilms, like you find in dental plaque, or wastewater stream infrastructure, et cetera.
In our Genemat system at Ginkgo, we have two chambers and the culture resides in one of them. I can transfer this culture to another refuge chamber so that I can sterilize and then rinse the principal vessel.
Meanwhile, the culture is safe in the other. And after we have restored the original conditions, I can transfer back. And then I can sterilize and rinse the second, refuge chamber. And I can complete a cycle by the end of which the probability of survival by sticking somewhere is absolutely zero. So the idea is that we just get rid of these biofilms and in a closed set up, which means I do not need to replace containers to open tubes or whatever, or to manually interact with the system. This paved the way for full automation of ALE in a fluidic setup that can dramatically limit the chances for contamination and system failure, and that can work 24/7 for as long as necessary.
What we achieved with the Genemat after several years of development is a standalone, autonomous fluidic apparatus that automates ALE, with tubes connecting growth chambers between themselves and to tanks containing growth media, sterilizing agents, water. This results in a closed circuit that is sterile, and there is no manual intervention required in it, and we can automate everything mainly through optical density measurement.
The achievement that we’ve done over the last 20 years was actually to have a system that works, that is really automated.
SA: How long can you run an ALE experiment for?
ST: Typically, the duration of an ALE campaign on the Genemat is around 3 months. That can be shorter or longer–it really depends on the project. The length of the experiment is no constraint to us, the Genemat can support ALE experiments for as long as necessary.
At CEA Genoscope, a French research center that co-owns the technology, an experiment has been running for about 10 years. I think that we have accumulated maybe 50,000 or 60,000 generations, you know, maybe 10 years which is maybe three or four times faster than doing the experiment by serial passaging.
The capacity of maintaining those cells growing in exponential phase always, or in different states, depending on what we want to do, it’s a pretty unique capacity. The Genemat will adjust the selective pressure to the actual adaptation of the microbes. And so we have a system that works 24/7 and we can reach our target faster.
Of course, it’s hard to imagine that a customer would come to us with a project that runs for 10 years! But our experiment shows the power of our ability to create a sterile environment and maintain cells for extended periods, while reaching the target faster.
SA: How does this tie into Ginkgo’s Foundry?
ST: We take samples from the Genemat during evolution that can be characterized at any point during the experiment. Historically, before we were acquired by Ginkgo, we had only been doing this work of inoculating the machine, evolving, taking samples, and shipping the samples to the customer, which also prevented us from having too much insight on the actual process and impact of the evolution going on in the Genemat.
But now, with Ginkgo’s Foundry, we can access the data generated with the samples. The ALE team continues having the same scope as we did previously, and we will ship those cryotubes through the Foundry, but we have access to the information of how the evolved strains performed now, and what were the paths taken by evolution. That’s very exciting for us. Depending on what’s needed for the project, we will isolate clones, perform basic characterization and dispatch to other Foundry services for phenotyping or genotyping.
SA: What’s the output of automated ALE after everything goes through the Foundry? Are you working with the entire population? A single clone?
ST: We collect samples from any experiment on a routine basis every week. That’s our standard. We do a basic QC and we store that in our freezers as a backup of evolution. And this happens for the duration of an experiment, typically a few months.
What we collect from our system is a polyclonal population. We collect them in a 1 ml cryotube so we can characterize them.
For this, usually we just first sequence a given number of clones to understand how many different genomes we have, so that we can further assess the phenotyping. And when we understand that, then we can test the different genotypic variants for how well they perform for the desired KPIs.
I like the way we do this: first sequence, understand how many genomes we have at this point in the ALE, and then we can then calibrate the characterization.
So usually the customer will get the best clones, generally, regardless of the nature of the program.
SA: What types of things would you consider in scoping Foundry services for a project?
ST: I would say a base scope of work includes a basic screening of the best clones against basic phenotypic indicators (KPIs). But if you want to have more insight on the performance, we might characterize other phenotypes using omics, fermentation–everything that Ginkgo can offer. Now, we can also sequence the strains to understand the beneficial mutations that we could use in a rational engineering campaign that might be running in parallel. At Ginkgo, this is something that could be done by the Systems Biology group.
So integrated into the Ginkgo’s Foundry, ALE is so much more powerful. Not only do you have the strains as an output, but now you can understand the pathway they took, how they perform, and have a roadmap for improving the phenotype in the background of your choice.
And the combination of these services, in one place without having to coordinate different efforts, that’s what makes it exciting to be a scientist at Ginkgo–you can understand a problem and find solutions. And that’s also an incredible service for our customers to have access to.
Read Part 1, Why ALE?, here
Read Part 3, Applications and Opportunities, here
 Simon Trancart joined Ginkgo through the acquisition of Altar, a French biotech company he co-founded and led as CEO. Altar specialized in automated adaptive laboratory evolution (ALE), a niche that Simon navigated with his background in engineering and civil engineering.
Simon Trancart joined Ginkgo through the acquisition of Altar, a French biotech company he co-founded and led as CEO. Altar specialized in automated adaptive laboratory evolution (ALE), a niche that Simon navigated with his background in engineering and civil engineering.
At Ginkgo, Simon leads the Adaptive Laboratory Evolution, based in Évry-Courcouronnes, France. Simon’s work focuses on the automated ALE process, which the performance of ALE campaigns. He has been instrumental in integrating the ALE team’s work with Ginkgo’s foundry services, enabling better execution and insight into ALE. Simon’s expertise extends to the application of ALE in various organisms and its coupling with rational design.
