Sneha Srikrishnan, Director of Growth, Business Development at Ginkgo Bioworks, is here to answer your burning questions around our new offerings in protein expression!
This conversation was moderated by Annick Saralegui, Senior Marketing, Growth Specialist at Ginkgo.
To kick things off, tell me, what does Ginkgo have to offer for customers developing proteins?
Sneha Srikrishnan: This is super exciting! So we have four different modules of work that we would like to repeatedly use to help our customers in protein expression wherever they are in the R&D cycle. The 4 include:
- Host evaluation: evaluate and compare your host strain with our suite of hosts
- Strain optimization: optimize your strain with genome modifications for better quality, functionality, and titer
- Classical strain improvement: use non-GM approaches to push titers higher
- Scale + fermentation: access our pilot and commercial scale fermentation facilities to increase production efficiency all in-house
For the first module, host evaluation, does this mean I can bring my own strain to Ginkgo? If I’ve spent a few long-and-hard years designing a host, can Ginkgo work with my strain?
Certainly! We can now engineer customer strains as much as we can engineer Ginkgo chassis strains. So that’s number one.
And what that means is, if our customers have strains that they really like and they enjoy using, bring it to us! We can improve on it and help them get to higher titers, provide better functionality, and even improve on process media optimizations or process development for those strains.
In the early days at Ginkgo, we were more comfortable just working with Ginkgo strain backgrounds and now we’ve opened doors to customer strain backgrounds. And why? Because we recognize that our customers have put a lot of time and effort into their R&D and their strain backgrounds and have invested in CapEx to build processes that go along with their strains. So instead of backtracking and eliminating the body of work that’s already been invested in, we can add to it.
Let’s say I’m not too sure what strain I should use, mine or Ginkgo’s. Can Ginkgo help me make the right call to reach my goals?
Yes. We can now perform host strain comparisons – early on.
That means if anyone is starting early on and doesn’t know whether to invest in a fungal strain background which is filamentous in nature, like a Trichoderma or an Aspergillus, versus say a Pichia we have, we’ll run in a host evaluation body of work, which is very quick.
It can be run within six weeks, maximum eight weeks, to provide a high level feedback on whether Pichia or the others have better better titers and are able to be a good match for being a production organism. So it gives you quick feedback and this can help with early stage GRAS filing.
Let’s say I’m a prospective customer and I’d like early access to a protein sample. Is this something Ginkgo offers?
Yes, that’s very new! If anybody wants protein samples to try and test for functionality during a strain engineering program, we’re willing to provide them with samples. These can be generated from ambr250 system fermentations or from 5L fermentations or even higher.
What I’ll also mention is Ginkgo has invested in building out our scaling facilities so that we can even run strains at a commercial scale. Why is this important for the protein world?
Under one roof, we have the ability of generating up to 10 grams of sample, if not at the kilogram scale when run at 3000 liters.
It can all be done in-house and this is very exciting because our partners can essentially receive samples, do some functionality testing, and if there’s anything to be changed from the same strain background to meet specs, we will now be able to fix the strain and not lose time in the process. So it’s time gained.
What’s so special about Ginkgo’s strain assets especially for, as an example, alternative food protein production?
I’m excited because we have developed cutting-edge strains for very special and complex, hard-to-engineer proteins such as iron-bound proteins. We’ve spent a lot of time determining how to modify the strains’ post-processing folding machinery in order to improve the titer and functionality of proteins.
So we’ve basically created a suite of strain backgrounds that can accommodate multi-copy integrations and codon optimization. We also have proprietary synthetic promoter systems that will allow for tunable and controllable expression which are not methanol-based. So methanol-free is based on simple carbon sources, which have very good COGS in terms of raw material usage.
We also have synthetic secretion signals for secreted protein. If you want to keep it intracellular, we can keep it intracellular and we’ve also developed a suite of strains which have unique helper genes or co-expression genes such as chaperones and protease knockouts.
Why is this important? Because we figured out ways in which the helper genes will help, post-translation, to package the proteins and help translate them across the ER, and then across the cell wall.
Essentially, we have identified unique ways to not just produce a protein but also to keep it stable once it’s secreted.
So all of this can be done in short spans of time. We have examples where we’ve run dairy-protein type projects. The first time we generated these strains, we invested close to 15 months worth of pre-work to generate the strains.
Because of the pre-work we invested, When we partnered with clients that needed iron-bound proteins, we were able to get from a single digit gram per liter, very quickly up to in the range of 5+ gram per liter scale for dairy protein within six months, which is significant time savings.
Can I test for functionality with Ginkgo?
We have some capability to test functionality within Ginkgo for a subset of samples. And if we are unable to run those functionality screens in-house, we have partnerships set up where we could be outsourcing to our partners for functionality screens for food proteins.
Assays such as iron binding can be done within Ginkgo.
Antimicrobial activity can be done within Ginkgo other more specific assays such as: melting temperatures, aggregation temperatures can also be done at Ginkgo. If there are more specific assays required, we can outsource to our partners.
I’m a prospective customer and have heard of EncapS and ALE. EncapS for Encapsulation & Screening and ALE for Adaptive Laboratory Evolution. Can you explain that a bit more and how I can leverage those services for my project?
The EncapS strategy would normally work if you have a strain that’s already making something. If you have a strain that’s making a product of interest or a protein of interest, and it’s making a very small amount of the protein and you want to get to multifold improvements very quickly, then go with EncapS. So with EncapS, what we can do is when you are already at reasonable titer in the gram per liter range, we can increase titers by 20, 30 percent or sometimes even reach 50 percent improvement.
We can run multiple rounds of classical mutagenesis or generate a library of semi-rational mutants. That semi-rational mutant library can then be quickly screened through encapsulated cells within nanoliter beads. And we can screen over 1 million clones in a single round.
Why is this useful? because when you screen for a million clones you can quickly get titer improvement jumps.
EncapS can also help you achieve better strain productivity.
We will essentially not just look for a high protein titer, but we’ll look for improved productivity. So we’ll directly read out for secreted protein, as well as the biomass.
Why is this good? Because you not only want to have high titer, you also want to have decent microbial growth. Otherwise your fermentation will be challenging. So if you want to have good productivity, you look for both good biomass and good protein titer. The EncapS method can help you look for both.
Now, where does ALE come into picture, if you want to switch carbon sources?
So let’s say that you have a strain that is going to grow on waste material and you want to circularize the process. You want to do waste valorization and you want to pick up C5 sugars in addition to C6 sugars. Then ALE is a great way to change the carbon preference or to open out the carbon preference as an example for the organism that’s already making something of your interest.
If you’re looking for change or improvement in the growth rate specifically then it can be considered. That’s how I would use it.
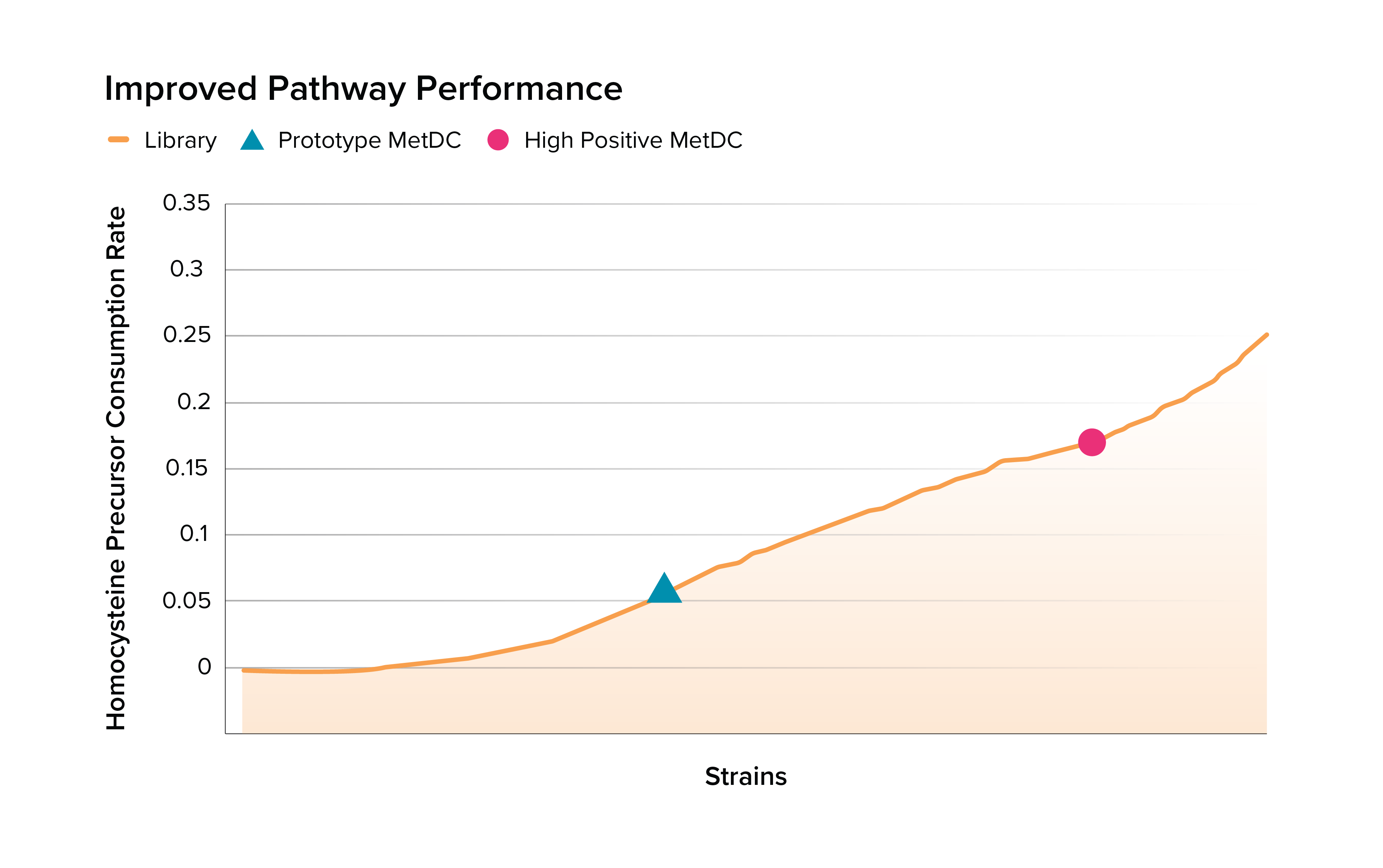
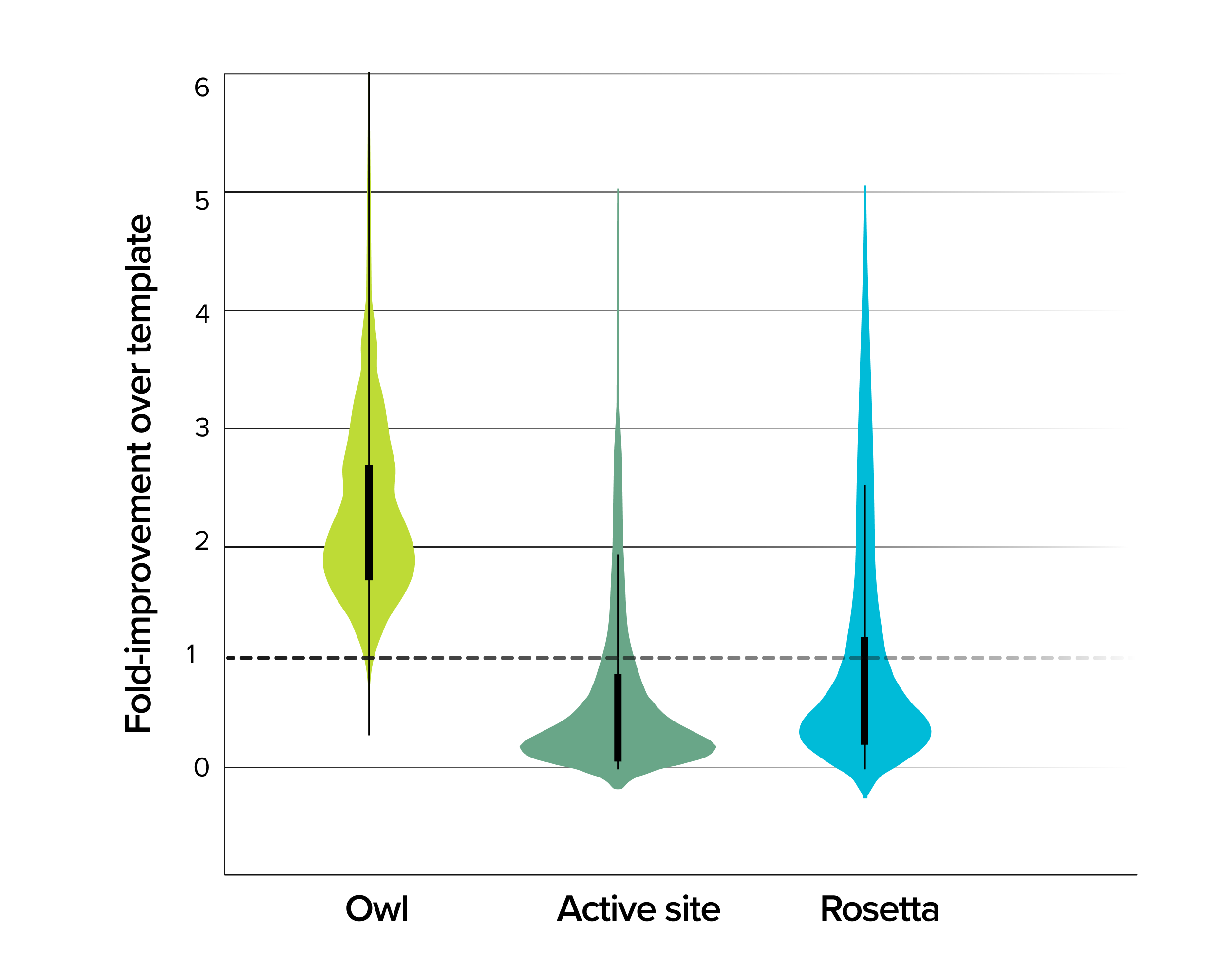
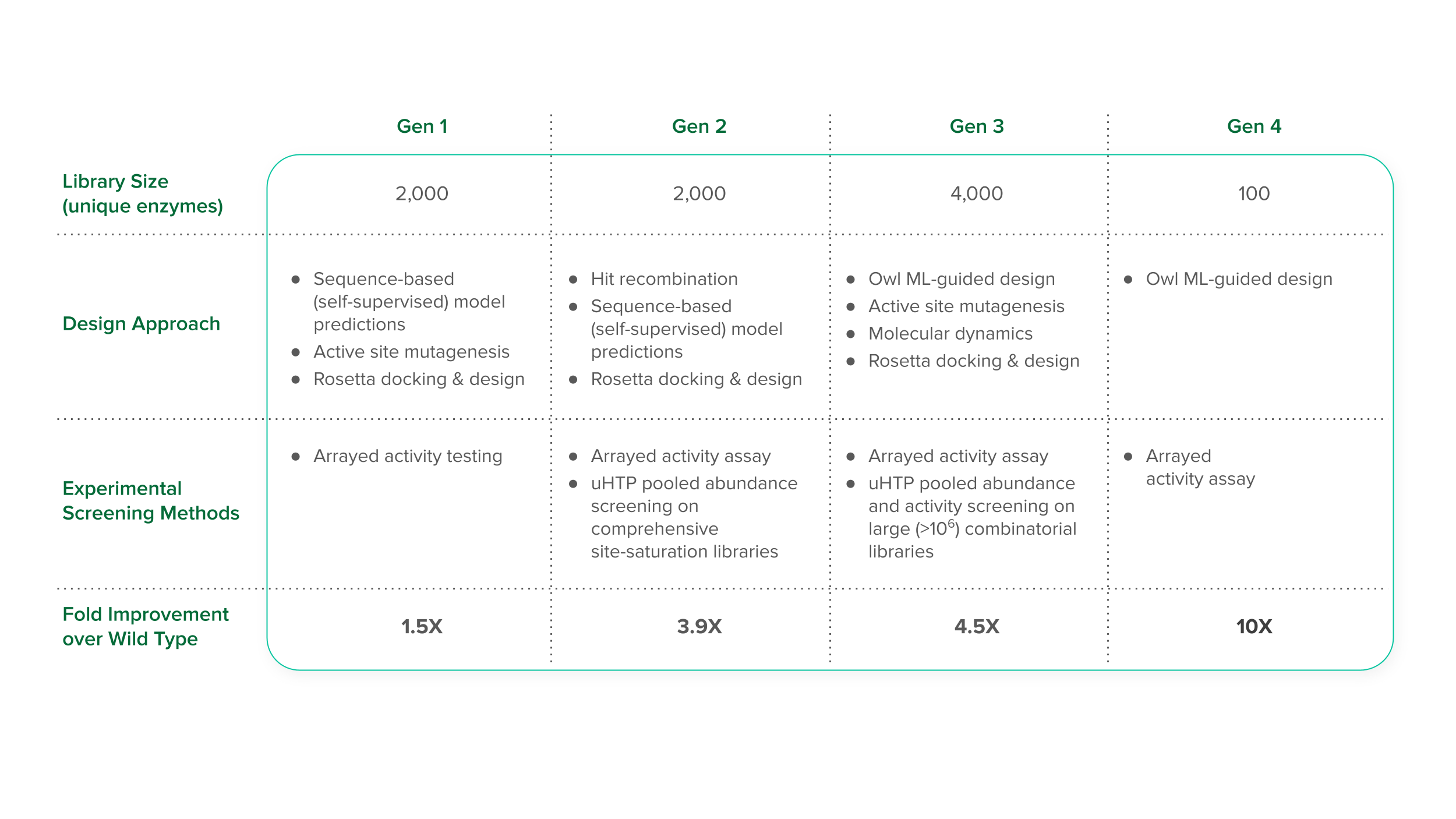
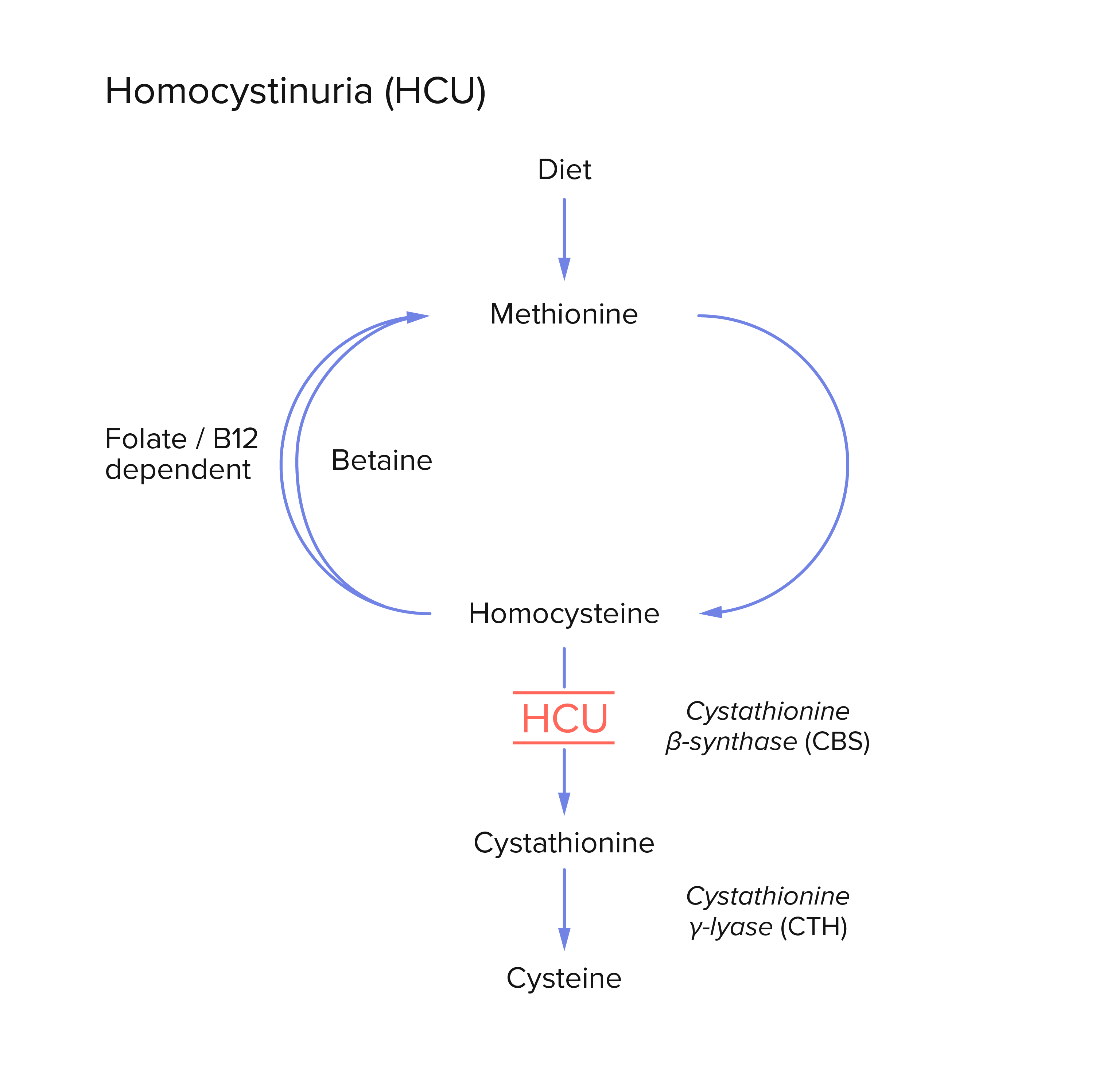 Solution | Boosting microbial metabolism for human disease
Solution | Boosting microbial metabolism for human disease